Atomic Modeling Language
4 posters
Page 1 of 2
Page 1 of 2 • 1, 2 
 Atomic Modeling Language
Atomic Modeling Language
AML - Atomic Modeling Language
The language of the nucleus. An expression of the microscopic world. Sculptures brought forth in all of their three dimensional greatness. Now you have that power at your disposal. What wonderful things will you create? Maybe you can unlock the mysteries of the elements.
AML is a language that allows you to create a nucleus, all the way down to the protons, neutrons and electrons. Yet it provides a simplicity that allows you to create structures with ease. While it is possible to define a proton stack by specifying every single proton, neutron and electron, you can also create a basic stack of the same size in 2 characters. This allows you to start with basic constructs until you have the main structure, making rapid changes easily.
There are 4 layers to the language. At the lowest level we have the building blocks of the elements: protons, neutrons and electrons. The lower-middle layer gives us proton stacks and neutron groups, which contain those particles. The next layer provides a way to link stacks together into a chain. Finally, the top layer allows us to join these together into a nucleus. Giving us various types of atoms.
Play with AML today at www.nevyns-lab.com/mathis/app/aml/aml.html!
Last edited by Nevyn on Fri Jan 04, 2019 9:02 am; edited 2 times in total
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Particles
There are 3 types of particles and they are all defined in the same way. A type prefix, optionally followed by an integer between 1 and 6 inclusive. If a number is not specified, it will default to 1.
<type><1-6>
Example:
P = 1 proton
P3 = 3 protons
N2 = 2 neutrons
E = 1 electron
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Particle Chains
We can link particles together into a chain in order to create proton stacks and neutron groups. There are limitations to how various particles relate to each other and you must be careful to get meaningful results.
In order to create a particle chain, you use the : operator to connect particles together. For example, if we wanted to create a proton stack with 2 neutrons between 2 protons, we connect them like this:
P:N2:P
The stack is arranged from bottom to top, so we start with a proton, then a level containing 2 neutrons, and a proton on top. Notice that we specified N2 to create a single layer of neutrons. If we used two distinct neutrons instead:
P:N:N:P
we would get a very different result. Each : operator creates a new level, so by separating the neutrons, 2 levels are created. Electrons are the same.
The proton does not work in the same way, though. A proton will always create a new level, even if declared with a number. Protons can only stack one way, but neutrons and electrons can be used in groups that arrange them into the plane orthogonal to the direction of the stack.
This does allow us to use some shortcuts when working with protons. Say we wanted to create 2 stacks like we created above. We could do it the long way:
P:N2:P:P:N2:P
or we could make it a little bit shorter by bring those middle protons, P:P, together into a P2 declaration:
P:N2:P2:N2:P
Later, we will see how to declare the same stack with only 2 characters.
This shorthand is more useful with odd-sized stacks, since we can group the protons without neutrons together:
P:N2:P3:N2:P
Electrons must be bound to a proton. At least one side of it must connect to a proton, but the other side may connect to any other particle, even another electron. It is possible to put electrons on the inside of a stack, even though this is generally not done. You will usually find them on the start and end of a stack declaration, such as this:
E:P:N2:P2:N2:P:E
but may find them inside on special occasions.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
The Stack
It is really good to be able to declare a proton stack with such precision, but most of the time, we don't really need to be so expressive. So the basic stack can be used to quickly declare a stack containing a specified number of protons. You do this by using the S type prefix, followed by the number of protons in the stack. You will also get an even number of neutrons, arranged so that the stack is built from as many P:N2:P constructs as possible.
S2
S5
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Nuclear Chains
Now that we can build a stack of particles, we can join them together by chaining a series of stacks. Each alternate stack is rotated 90° so that one emits into the pole of the other. The - operator is used between stacks to link them into a nuclear chain.
S6-S4-S2
S6-P:N2:P3:N2:P-S3
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Atoms
We now have enough constructs to create an atom. All we need is to impose a little structure using the building blocks that we have. We start with the core of an atom. This is a single proton stack which can be declared using a basic stack or a constructed stack of particles. Around this core stack, we can arrange up to 6 nuclear chains, all pointing away from the core stack.
The first 2 chains can be put onto the top and bottom of the core. This arranges the chains such that they emit into the core. We call these the axial level.
The next 4 chains can be placed around the core and are called the carousel level. The first stack in a carousel chain is receiving charge from the core of the atom.
To declare an atom, we start with the core stack:
S4
then we add in pillars and caps:
S4 <Axial S4-S4, S4-S4>
this introduces the level operators, < and >. A level must specify its type as the first token inside of the openning token. The above AML expression uses the full Axial keyword, but just A is enough. Similarly, you can create carousel chains by using the Carousel or C keywords.
S4 <Axial S4-S4, S4-S4> <Carousel S4, S4, S4, S4>
There are 3 more level types that can be used that allow you to specify the dimension on which 1 or 2 nuclear chains can be placed. Use X, Y, or Z
S4 <Y S4-S4, S4-S4> <X S4, S4> <Z S4, S4>
Each atomic level, Axial and Carousel or X, Y and Z, has a comma separated list of nuclear chains. All of them can accept 2 chains, but the Carousel level can accept 4. The order of the nuclear chains is positive then negative. For the Carousel level, it is +X, -X, +Z, -Z, and is equivalent to <X +, -> <Z +, ->. So the Axial or Y levels can set the North and South positions, while the Carousel can set the East, West, Front and Back positions. The X level sets the East and West while the Z level sets the Front and Back.
Last edited by Nevyn on Fri Jan 04, 2019 7:32 pm; edited 2 times in total (Reason for editing : Added a description of nuclear chain ordering within a level declaration)
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Getting Attached
Some elements need to place some stacks that are bonded to the pillar stacks, which are the first stacks in each chain of the axial level. These can be placed after the stack that they are bonded to using the attachment operators [ and ].
S4 <Axial S4[P]-S4-S1, S4[P]-S4-S2> <Carousel S4-S2, S4-S2, S4-S1, S4-S1>
Each attachment can contain 1 or 2 stacks with the first one placed in the positive X in the north and negative X in the south. The second stack is placed on the other side, so negative X in the north and postive X in the south. If you don't want a stack in the first position but do in the second, then use the null operator _ in the first position.
S4 <Axial S4[_,P]-S4-S1, S4[_,P]-S4-S2> <Carousel S4-S2, S4-S2, S4-S1, S4-S1>
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
.

Wow Nevyn, you’ve created a simple to use and understand atomic code.
Putting on my critical cap, there I was following along nicely, totally engrossed - and you stopped. Where’s the Congratulations! You’ve created atomic number 47 - Silver, aka Ag. Please consider adding a help page containing these same instructions; and a link back to your lab. Meanwhile, back at the Lab, where is the Atomic Modeling Language (aml) linked? A little added confusion, this app is not the same as the Atomic Model Editor – which displays the specific elements the User selects. It’s getting difficult telling the apps apart. Please consider some renaming or explaining a bit of the bigger picture, directing the reader to the Atomic Viewer or Molecular Bonding Language next.
https://www.nevyns-lab.com/
Good Stuff Nevyn.
.

Wow Nevyn, you’ve created a simple to use and understand atomic code.
Putting on my critical cap, there I was following along nicely, totally engrossed - and you stopped. Where’s the Congratulations! You’ve created atomic number 47 - Silver, aka Ag. Please consider adding a help page containing these same instructions; and a link back to your lab. Meanwhile, back at the Lab, where is the Atomic Modeling Language (aml) linked? A little added confusion, this app is not the same as the Atomic Model Editor – which displays the specific elements the User selects. It’s getting difficult telling the apps apart. Please consider some renaming or explaining a bit of the bigger picture, directing the reader to the Atomic Viewer or Molecular Bonding Language next.
https://www.nevyns-lab.com/
Good Stuff Nevyn.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Thanks, Airman, I'm pretty happy with how it came out. I based it on the MBL code and just cut out and reshaped it for this new purpose. So it came together quite quickly. I plan on adding a specification and guide, just like MBL has. There is no link on my home page because it isn't officially released yet. I like to see what you guys think before I do that. There is also some more work to do on the larger elements, like Uranium, that contain multiple nuclei. I have the language parts defined, but haven't written it into the parser or visualizer.
AML has no connection to Atomic Model Editor (also called Atomic Viewer, or AV), even though MBL does use code from AV to render the atoms. This is a completely new rendering engine, but I have used the same styles such as colors and textures, so things do look a lot alike. There are some small differences though, mostly in the placement of particles in a stack.
I'm still amazed at how much you can do with AML in such a small amount of text. Take a look at Silver:
S4<A S4[P]-S4-S1,S4[P]-S4-S2><C S4-S2,S4-S2,S4-S1,S4-S1>
In its smallest expression, we only need 56 characters to define it. That's a pretty big nucleus. The equivalent AV model uses about 2350 characters over 127 lines. That's a 42:1 compression ratio. Gold would probably be about the same length. And it is just so easy to manipulate. I love it!
AML has no connection to Atomic Model Editor (also called Atomic Viewer, or AV), even though MBL does use code from AV to render the atoms. This is a completely new rendering engine, but I have used the same styles such as colors and textures, so things do look a lot alike. There are some small differences though, mostly in the placement of particles in a stack.
I'm still amazed at how much you can do with AML in such a small amount of text. Take a look at Silver:
S4<A S4[P]-S4-S1,S4[P]-S4-S2><C S4-S2,S4-S2,S4-S1,S4-S1>
In its smallest expression, we only need 56 characters to define it. That's a pretty big nucleus. The equivalent AV model uses about 2350 characters over 127 lines. That's a 42:1 compression ratio. Gold would probably be about the same length. And it is just so easy to manipulate. I love it!
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
I made a slight update to handle neutrons better. If there are multiple neutrons in a single level, such as N2, then they will work as they did before and not take up much space between the bounding protons. However, if there is only a single neutron, then it will take up as much space as a proton would. It is also placed at the center like a proton.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
.

Oops, correction. This, rather than the previous image, is the current default output for Silver's AML.
I changed the view while I was looking it over before finally taking its image. That's unfortunate in that I rather like the current default image - looking directly at Ag from either its X or Z directions.
.

Oops, correction. This, rather than the previous image, is the current default output for Silver's AML.
I changed the view while I was looking it over before finally taking its image. That's unfortunate in that I rather like the current default image - looking directly at Ag from either its X or Z directions.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Multi-Nuclei Atoms
The largest of atoms are not really atoms, and could be thought of as molecules. However, they are on the periodic table, so we continue to call them atoms. Therefore, AML must support them. Since the nuclear structure is so different from the rest of the elements, we must treat them as a special entity. This introduces a fifth level to our language.
What we need to do is define multiple atoms and bond them together. So we do just that, separating each atom with the nuclei operator ~.
S4<Y S4-S4-S1,S4-S4-S2> ~ S6<Y S6-S4-S1,S6-S4-S2>
But that didn't really do what we wanted now, did it? Sure, we have 2 nuclei in the same atom, but they aren't exactly bonded. They also don't bond in the right places. We want the 2 single proton hook stacks to come together. We could accomplish that by changing one of the nuclei declarations to swap the single and double hook stacks around. Wouldn't it be a lot better if we could just flip one of those nuclei over?
S4<Y S4-S4-S1,S4-S4-S2> ~ !S6<Y S6-S4-S1,S6-S4-S2>
We use the flip operator ! before the nuclei that we want to rotate by 180°. This allows us to keep the same nuclei definition used in any situation, and just flip it over when we want to.
However, we still don't really have the structure that we want. Now we have the correct hook stacks bonding, except that, well, they aren't bonding. To solve this, we could alter both of the nuclei and take a single proton hook stack from one and put it into the other as a double proton stack. That would work and look fine, but there is a better way.
What we can do, is take the single hook stacks from both nuclei, put them together into a double stack and make it a new nuclei that sits between the others.
S4<Y S4-S4,S4-S4-S2> ~ P2 ~ !S6<Y S6-S4,S6-S4-S2>
What have we done? That ruined everything! The middle stack is not in the correct position to bond with the other nuclei, and we can't flip it. What can we do? Easy, we just use the rotate operator ^ before the atom that we want to rotate by 90°
S4<Y S4-S4,S4-S4-S2> ~ ^P2 ~ !S6<Y S6-S4,S6-S4-S2>
Voila! We have a beautiful, multi-nuclei atom. The good thing about splitting this into 3 nuclei, is that we can easily manipulate the bonding nucleus. Say we wanted to put neutrons on either side of that nucleus, then we just add them logically:
S4<Y S4-S4,S4-S4-S2> ~ ^N:P2:N ~ !S6<Y S6-S4,S6-S4-S2>
If we want to be more precise than that, which we might for purposes of calculating data from these expressions, then we could add those neutrons to the axial level like this:
S4<Y S4-S4,S4-S4-S2> ~ ^P2 <A N,N> ~ !S6<Y S6-S4,S6-S4-S2>
How cool is that?
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
.
Very cool, very efficient, easy to use and understand. Atomic Viewer shows the elements, all well and good; but in my opinion, this particular application - Atomic Modeling Language (AML) - provides an excellent - the best yet - means of learning charged field atomic structures.
Start at the center stack and build in six directions: up and down the axial; and out the +/-X and +/-Z for the carousal. Almost as simple as counting from the center nucleus. The flip and rotate options extend functionality. You already have the makings of an atomic structures tutorial above. I expect you to show us how to build angled bonds or molecular rings at any moment.
Those multi-nucleus elements are rather long, obviously you'll need to rotate the view of the Y axis by 90 degrees as Miles and you have done in displaying these longer elements in the interactive papers.
Consider adding a proton counter somewhere.
.
Very cool, very efficient, easy to use and understand. Atomic Viewer shows the elements, all well and good; but in my opinion, this particular application - Atomic Modeling Language (AML) - provides an excellent - the best yet - means of learning charged field atomic structures.
Start at the center stack and build in six directions: up and down the axial; and out the +/-X and +/-Z for the carousal. Almost as simple as counting from the center nucleus. The flip and rotate options extend functionality. You already have the makings of an atomic structures tutorial above. I expect you to show us how to build angled bonds or molecular rings at any moment.
Those multi-nucleus elements are rather long, obviously you'll need to rotate the view of the Y axis by 90 degrees as Miles and you have done in displaying these longer elements in the interactive papers.
Consider adding a proton counter somewhere.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Oooohhh, a panel showing calculated statistics! I like it. I wasn't even thinking in that direction.
The atoms are generally oriented N to S, so I was thinking about how to change the rendering area into a more vertical orientation. It doesn't work well with the controls though. I may have to rethink how they are presented.
A way to orient the whole atom along the main dimensions may work though. I actually removed that from the code base as it came from MBL. I didn't want to have to deal with those direction issues again. I can do it a different way this time though. I don't have the same problems that I did with MBL.
I just added motion to the electrons and neutrons. They spin around the stack axis. This sets the ground work for the emission shaders. They'll be easy to add in now.
The atoms are generally oriented N to S, so I was thinking about how to change the rendering area into a more vertical orientation. It doesn't work well with the controls though. I may have to rethink how they are presented.
A way to orient the whole atom along the main dimensions may work though. I actually removed that from the code base as it came from MBL. I didn't want to have to deal with those direction issues again. I can do it a different way this time though. I don't have the same problems that I did with MBL.
I just added motion to the electrons and neutrons. They spin around the stack axis. This sets the ground work for the emission shaders. They'll be easy to add in now.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Multiplication
You can use multiplication groups to expand a given section, but you must be careful what you are multiplying. You can only use the multiplication mechanism within a particle chain or a nuclear chain. The multiplication group can not start in one and end in the other. It must be completely contained within either type of chain. However, the multiplication group may start in a nuclear chain and contain a complete particle chain, as long as it ends in the nuclear chain.
A multiplication group starts with a ( and ends with a ) followed by a positive integer.
Multiplication occurs at the token level. You are multiplying the exact characters that are inside of the group. Therefore, you must make sure that it contains the necessary linking operators : or -.
Multiplying with a Particle Chain
Let's take a standard proton stack containing 2 neutrons between 2 protons.
P:N2:P
If we wanted to use 3 of these to create a 6 proton stack, we could use a multiplication group like this:
(P:N2:P)3
If we wanted to add electrons to both poles, we just add them outside of the multiplication group:
E:(P:N2:P)3:E
Multiplying a Nuclear Chain
We can do the same things with a nuclear chain.
S4 <A (S4-)3>
S4 <A (P:N2:P-)2-S2>
Notice that we have to put a - inside of the multiplication group so that everything makes sense once it is expanded. You may have to do the same with the particle chains and the : operator, but they seem to be more resilient. The nuclear chains are not and require those linking tokens. Strictly speaking the link token after the multiplication group is not required, bit it doesn't hurt if used and makes it more readable.
We can actually nest a particle multiplication inside of a nuclear chain multiplication group.
S4 <A ((P:N2:P)2-)2-S2>
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
This is soooo awesome, Nevyn! Clean and simple, too. I'm diggin' it. It's amazing how rapidly you put things together once you get going! Well at least it is, from my perspective. I stumble along and hit roadblocks right and left in my software, and you guys just power around them by writing your own stuff. It's rather impressive, but even moreso because of the topic we're all working on. I feel like this is a LOT of progress!
Gonna play with it awhile and see how it goes! And yeah, you should definitely plop this onto your site once you get time and can compile the help. It's great.
Gonna play with it awhile and see how it goes! And yeah, you should definitely plop this onto your site once you get time and can compile the help. It's great.
Jared Magneson- Posts : 525
Join date : 2016-10-11
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Element Type
I have added a new field to the Element Information panel that attempts to guess what type of atom you have created. It sets it to Noble, Conductive or Magnetic. If it can't figure it out, it sets it to Unknown. This probably needs more work, but it seems to work well at the moment.
The type is determined by looking at the arrangement of protons and stacks. A conductive element has a difference in the axial hook stacks. A magnetic element has a balance, but must also have a carousel level. A noble element does not have hook stacks and must be balanced all round.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
My suite of charge shaders have been added to AML, and I even created a new one that represents the output of a charge channel. Basically the opposite of the intake shader.
Charge moves in defined directions through the nucleus. The axial level is an input and the carousel is an output.
There is a new settings dialog that you can access with the options button (...). Some of these will be reconfigured soon, as they have changed a bit recently and some of them are redundant. They allow you to turn the charge shaders on/off.
Charge moves in defined directions through the nucleus. The axial level is an input and the carousel is an output.
There is a new settings dialog that you can access with the options button (...). Some of these will be reconfigured soon, as they have changed a bit recently and some of them are redundant. They allow you to turn the charge shaders on/off.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
.

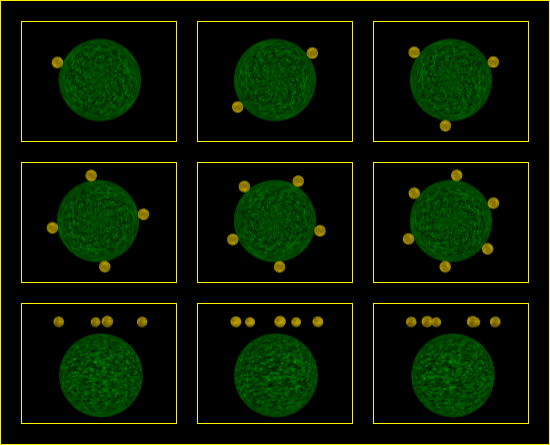
Single layer neutron configurations N1 thru N7.
Great progress. As if you don't already know, I'm here to help. I said a tutorial would be a good thing, knowing that that would require additional work, so I made these two images for convenient reference. Feel free to request any changes, additional images or taskings.

Proton stacks - each proton is a layer - P1 thru P7.
.

Single layer neutron configurations N1 thru N7.
Great progress. As if you don't already know, I'm here to help. I said a tutorial would be a good thing, knowing that that would require additional work, so I made these two images for convenient reference. Feel free to request any changes, additional images or taskings.

Proton stacks - each proton is a layer - P1 thru P7.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
I think it will need a few charts. The neutron groups is a good one. Simple stacks, like you have already done. Then the standard stacks, S1 - S6. Unfortunately you can't generate a single chain of stacks. You must specify a core stack for the atom and then you can put chains on to that. Some of my links above don't actually work because of that. I think at some point you could do it, but then once I started to deal with atoms, it disappeared. Maybe I could have some special character that tells the parser to treat it as a stack, rather than an atom. while you don't need it for actually building elements, it is quite helpful for these types of tutorials and charts. I guess you could generate them with the core, and then crop the core out in an image editor. That might work, but I didn't become a software developer to do things the hard way!
I would turn off all charge shaders for any images that are not specifically there to show charge. Keeps it cleaner and easier to understand.
Another good image would contain 2 2-proton stacks next to each other. The first one does not have neutrons (P:P) but the second one does (P:N2:P). This will show the size differences. I will probably have a section dedicated to building a stack, particle by particle.
I would turn off all charge shaders for any images that are not specifically there to show charge. Keeps it cleaner and easier to understand.
Another good image would contain 2 2-proton stacks next to each other. The first one does not have neutrons (P:P) but the second one does (P:N2:P). This will show the size differences. I will probably have a section dedicated to building a stack, particle by particle.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
So I finally got a chance to look into that stack size bug, and found that it is already correctly limiting the default stacks (S1 to S6), but it does not limit a declared stack (P:P) and I am not sure if it should. After all, it is what the user asked for. I might just leave it as-is. You can already build invalid structures in other ways. I don't think the language needs to impose all of the constraints of the theory, only provide the freedom to express that theory correctly.
A standard stack is declared with the S prefix, S1 to S6, and is built using as many P:N2:P blocks as possible. The stacks you have created do not have any neutrons in them, and that is good, I want them that way for the initial parts of the tute, but the next part wants those neutrons in there.
Remove P7 from the stack image. We only ever need 1 to 6 of any particle.
A standard stack is declared with the S prefix, S1 to S6, and is built using as many P:N2:P blocks as possible. The stacks you have created do not have any neutrons in them, and that is good, I want them that way for the initial parts of the tute, but the next part wants those neutrons in there.
Remove P7 from the stack image. We only ever need 1 to 6 of any particle.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Thanks, Airman, that's great.
Notice how the protons that have neutrons between them have a larger gap than those that do not? I'm wondering whether or not I should make that equal. It isn't really a problem, apart from S3, because the lone proton sits a little closer to the others and this breaks the symmetry.
Notice how the protons that have neutrons between them have a larger gap than those that do not? I'm wondering whether or not I should make that equal. It isn't really a problem, apart from S3, because the lone proton sits a little closer to the others and this breaks the symmetry.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
.
Bug report. Entering the following two AML codes results in an erroneous display of just S6.
S6-S4-S2 // Only S6 is displayed
S6-P:N2:P3:N2:P-S3 // Only S6 is displayed
///////////////////////
Going over your notes, do you see any value in creating the following table:
<1-6>
List all types: E, N, P, S
E electron, N neutron(s), P proton(s), S includes p:n2:p groups
Example:
P = 1 proton
P3 = 3 protons
N2 = 2 neutrons
E = 1 electron
<->
List all operators: -
' - ' 90 deg link, ' : ' creates a new level, ' < ' begin, ' > ' end,
' X ' left/right, ' Y ' North/ south, ' Z ' ' Axial or A '
.
I don’t see a problem. The lack of symmetry between the three protons in S3 may be due to the Neutrons, I wouldn't expect the protons to space 'perfectly' with the neutrons' present.It isn't really a problem, apart from S3, because the lone proton sits a little closer to the others and this breaks the symmetry.
Bug report. Entering the following two AML codes results in an erroneous display of just S6.
S6-S4-S2 // Only S6 is displayed
S6-P:N2:P3:N2:P-S3 // Only S6 is displayed
///////////////////////
Going over your notes, do you see any value in creating the following table:
List all types: E, N, P, S
E electron, N neutron(s), P proton(s), S includes p:n2:p groups
Example:
P = 1 proton
P3 = 3 protons
N2 = 2 neutrons
E = 1 electron
List all operators: -
' - ' 90 deg link, ' : ' creates a new level, ' < ' begin, ' > ' end,
' X ' left/right, ' Y ' North/ south, ' Z ' ' Axial or A '
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
That bug is what I was eluding to earlier. You can't enter in just a nuclear chain, because it is trying to interpret it as an atom. So the first stack is added as the core of the atom, but the rest gets chopped off (well, it is ignored). That's why I thought about adding a special character to the start of the expression that tells the parser to treat it as a nuclear chain, rather than an atom.
A picture of each particle would be good. Just proton, neutron and electron. You could do an image with electron groups, as you have the neutrons. It will be the same, but it may be worth using.
Apart from that, it is a bit hard to tell what I need at the moment. You could get an image from each link I have used in the descriptions above (only those with the special headings at the start of the post). I imagine that the posts above will be pretty close to the tutorial/specification.
A picture of each particle would be good. Just proton, neutron and electron. You could do an image with electron groups, as you have the neutrons. It will be the same, but it may be worth using.
Apart from that, it is a bit hard to tell what I need at the moment. You could get an image from each link I have used in the descriptions above (only those with the special headings at the start of the post). I imagine that the posts above will be pretty close to the tutorial/specification.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Hmm, it seems you can't have just electrons in an expression. I was thinking of exactly what you have for the neutrons, but with electrons, but I just tried it, and it generates an error. That's fine though. I can work without them. I might just use an image of a proton stack with electrons on either side. Something like E:P:N2:P:E.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
It is probably better to use a proton than a neutron, since you are supposed to attach electrons to protons. I'm not sure what I will use at this point, but those images do look cool. The proton disc makes a good background.
I have created the basis of the spec, just by copying these posts into a HTML document and adding in some formatting. I haven't put any images in there yet, but have the placeholders in there, they just don't show anything because the images don't exist. It feels a bit more like a tutorial than a spec, so I might end up using it for that and create a more formal specification page. AML is a bit more complicated than MBL in some regards, since it has multiple levels, so a bit more explanation is needed. So maybe the current version will be fine. I'll try to add a bit more detail into some sections and see how it goes.
I have created the basis of the spec, just by copying these posts into a HTML document and adding in some formatting. I haven't put any images in there yet, but have the placeholders in there, they just don't show anything because the images don't exist. It feels a bit more like a tutorial than a spec, so I might end up using it for that and create a more formal specification page. AML is a bit more complicated than MBL in some regards, since it has multiple levels, so a bit more explanation is needed. So maybe the current version will be fine. I'll try to add a bit more detail into some sections and see how it goes.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Really Cool!
I've been playing with it since last weekend. Ace work Nevyn and great debugging LTAM and Jared. This is a little more flexible in many ways than the Periodic table. If there was a decent configurable RestApi we could throw an Extension (home-built) from KNIME at it for datamining.
www.knime.org

I've been playing with it since last weekend. Ace work Nevyn and great debugging LTAM and Jared. This is a little more flexible in many ways than the Periodic table. If there was a decent configurable RestApi we could throw an Extension (home-built) from KNIME at it for datamining.
www.knime.org
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
What would the REST API need to do? What does Knime do? I had a quick look at it, but I'm still not sure exactly what it would do for me if I used it. I guess your thinking of the machine learning aspect. That could be an interesting project. If you have any knowledge of that sort of stuff (cause I have very little), then maybe you could give an outline of what you would want it to do. I might be able to see how to help you then.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
.
Nevyn, please excuse my efforts at ‘help’. I'm sure I've given you the perceived slight or two - unintentionally, of course. I’m a team player, getting the charge field out there is the primary goal.
With respect to the colored glossies, adding section images is easy as each section contains plenty of AML code to choose from. I can’t really help any further until you specify a few formatting details. For example, all the images I've shown are draft suggestions - none are final. They cannot be final until the desired images are recreated in the desired size.

Incremental improvement (?) (square panes) to the Neutron set, N1 thru N6 images.
Another, note N5 in the square pane Neutron set isn’t very accurate – the direction of the pentagon is a little off, in fact none of the polygons are oriented perfectly. I think they’re Ok for real screen captures of a rotating configuration with luck, Paint and my less than perfect eyeball, but it isn’t my decision. They can all easily be made more precise by adding an AutoCAD rotation - at no additional cost, if you wish.
A spec HTML document! That sounds promising, what does it do? You make it sound reviewable, so I tried opening https://www.nevyns-lab.com/mathis/app/aml/aml.html in Inspector Mode. That’s not something I normally do since I don’t know what the heck I’m doing. I see plenty of links to many source files – several of which I’m familiar, but none – as far as I tell, are openable or reviewable.
Hey Cr6, I think AML is a proton/neutron/electron construction set that makes atoms and proton matter easier to understand. At the other end from understanding, Data Science and Machine Learning are well beyond my ken. Are you suggesting we give machines Nevyn's code, and let them start building proton matter structures?
.
Nevyn, please excuse my efforts at ‘help’. I'm sure I've given you the perceived slight or two - unintentionally, of course. I’m a team player, getting the charge field out there is the primary goal.
With respect to the colored glossies, adding section images is easy as each section contains plenty of AML code to choose from. I can’t really help any further until you specify a few formatting details. For example, all the images I've shown are draft suggestions - none are final. They cannot be final until the desired images are recreated in the desired size.

Incremental improvement (?) (square panes) to the Neutron set, N1 thru N6 images.
Another, note N5 in the square pane Neutron set isn’t very accurate – the direction of the pentagon is a little off, in fact none of the polygons are oriented perfectly. I think they’re Ok for real screen captures of a rotating configuration with luck, Paint and my less than perfect eyeball, but it isn’t my decision. They can all easily be made more precise by adding an AutoCAD rotation - at no additional cost, if you wish.
I have created the basis of the spec, just by copying these posts into a HTML document and adding in some formatting. I haven't put any images in there yet, but have the placeholders in there, they just don't show anything because the images don't exist. It feels a bit more like a tutorial than a spec, so I might end up using it for that and create a more formal specification page. AML is a bit more complicated than MBL in some regards, since it has multiple levels, so a bit more explanation is needed. So maybe the current version will be fine. I'll try to add a bit more detail into some sections and see how it goes.
A spec HTML document! That sounds promising, what does it do? You make it sound reviewable, so I tried opening https://www.nevyns-lab.com/mathis/app/aml/aml.html in Inspector Mode. That’s not something I normally do since I don’t know what the heck I’m doing. I see plenty of links to many source files – several of which I’m familiar, but none – as far as I tell, are openable or reviewable.
Really Cool!
I've been playing with it since last weekend. Ace work Nevyn and great debugging LTAM and Jared. This is a little more flexible in many ways than the Periodic table. If there was a decent configurable RestApi we could throw an Extension (home-built) from KNIME at it for datamining.
www.knime.org
Hey Cr6, I think AML is a proton/neutron/electron construction set that makes atoms and proton matter easier to understand. At the other end from understanding, Data Science and Machine Learning are well beyond my ken. Are you suggesting we give machines Nevyn's code, and let them start building proton matter structures?
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
No slights perceived by me, and if I did, I would just assume it was not intended as such and try to interpret it a different way.
The positions of the neutron groups does not worry me. If you look closely, you will notice that the upper-left neutron is always in the same position. That is the starting position and the rest are arranged at set angles from that. I did choose that position for some reason, maybe just because I felt like it at the time, but I think there was something but I can't remember. Maybe it just looked better. Actually, I think it was to keep the neutrons away from other things, such as through-charge streams. We really only need 1 to 3, as Miles has never used more than 3 neutrons in a group. Those 3 arrangements work quite well, so I'm not worried about 4 to 6. If it bothers you, and I know that desire for symmetry well, then go ahead and rotate them. As long as it shows the right number of neutrons, then I am happy.
I haven't uploaded the spec yet, because it is not ready. If it was, and you can even do this now, you can just remove the filename from the URL to get to the parent page: www.nevyns-lab.com/mathis/app/aml. That will contain links to the rest of the pages.
Note that the Inspector mode, or browser debugger, will only show you the files that the current page has downloaded. It will not show you the contents of the directory on the server, which you would need to see other pages.
I didn't do much work on AML last night. I did make some changes as I wanted to add a setting to stop the rotation of neutrons and electrons. This makes it much easier to capture an image and know what it will be. At the moment, it relies on when you press the capture button in the image dialog. I got that working, but didn't upload the changes.
That is because I got side-tracked on a new project that I will present soon. Another language, can anyone guess what that might be for?
The positions of the neutron groups does not worry me. If you look closely, you will notice that the upper-left neutron is always in the same position. That is the starting position and the rest are arranged at set angles from that. I did choose that position for some reason, maybe just because I felt like it at the time, but I think there was something but I can't remember. Maybe it just looked better. Actually, I think it was to keep the neutrons away from other things, such as through-charge streams. We really only need 1 to 3, as Miles has never used more than 3 neutrons in a group. Those 3 arrangements work quite well, so I'm not worried about 4 to 6. If it bothers you, and I know that desire for symmetry well, then go ahead and rotate them. As long as it shows the right number of neutrons, then I am happy.
I haven't uploaded the spec yet, because it is not ready. If it was, and you can even do this now, you can just remove the filename from the URL to get to the parent page: www.nevyns-lab.com/mathis/app/aml. That will contain links to the rest of the pages.
Note that the Inspector mode, or browser debugger, will only show you the files that the current page has downloaded. It will not show you the contents of the directory on the server, which you would need to see other pages.
I didn't do much work on AML last night. I did make some changes as I wanted to add a setting to stop the rotation of neutrons and electrons. This makes it much easier to capture an image and know what it will be. At the moment, it relies on when you press the capture button in the image dialog. I got that working, but didn't upload the changes.
That is because I got side-tracked on a new project that I will present soon. Another language, can anyone guess what that might be for?
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Airman, I forgot to mention the sizes of the images. It really depends on where they are used in the HTML document. The neutron group image looks about the right size for that content. Most images of a single AML expression will need enough size to show the element clearly. Maybe try exporting them at 480 and see how that goes. You might even be able to export them at 720 and then crop out a lot of the black areas.
Or, we use quite large images and I scale them down on the page, but each image is a link to the full sized version. If you find any that really want to be large, then don't hold back, just take them as you need to.
While this is quite dry content, see if you can get a bit creative and make the atoms look good. Sometimes the straight-on perspective is fine, other ones might want a bit of rotation to the atom, letting the insides become more visible.
It might be best just to get something done, then I can put them into the page and upload it. That will give you more of an idea of where the images are being used and you might see a better way to do some of them. Nothing is set in stone just yet. There is plenty of room for creativity, but we don't need to reach that level on the first attempt.
Really, anything you do is helpful and appreciated.
Or, we use quite large images and I scale them down on the page, but each image is a link to the full sized version. If you find any that really want to be large, then don't hold back, just take them as you need to.
While this is quite dry content, see if you can get a bit creative and make the atoms look good. Sometimes the straight-on perspective is fine, other ones might want a bit of rotation to the atom, letting the insides become more visible.
It might be best just to get something done, then I can put them into the page and upload it. That will give you more of an idea of where the images are being used and you might see a better way to do some of them. Nothing is set in stone just yet. There is plenty of room for creativity, but we don't need to reach that level on the first attempt.
Really, anything you do is helpful and appreciated.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
.
A little more to chose from:

Another possible Particle view of one through six electrons. E:P:N2:P:E1 - E6.

Boron. An example of a particle chain.
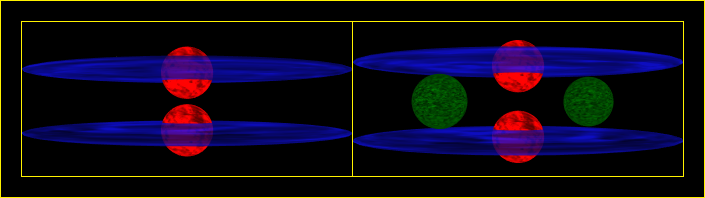
Improved "comparing the vertical difference between P2 and S2" option 1.

Improved "comparing the vertical difference between P2 and S2" option 2.
I'm happy with the settings page. The image saver works fine. My only gripe is, as previously mentioned, if we are limited by the view's vertical dimension, how can we reorient the desired image 90deg in order to fit the subject into the wider (longer) view dimension?
Funny, I've mentioned always using the print screen button - and finishing up in Paint. By some divine co-inky-dink, last night I received notice that Paint will shortly be moving to the Microsoft store, where it will be made available for free. I suppose it will no longer be bundled with Microsoft products. Time marches on.
.
A little more to chose from:

Another possible Particle view of one through six electrons. E:P:N2:P:E1 - E6.

Boron. An example of a particle chain.

Improved "comparing the vertical difference between P2 and S2" option 1.

Improved "comparing the vertical difference between P2 and S2" option 2.
I'm happy with the settings page. The image saver works fine. My only gripe is, as previously mentioned, if we are limited by the view's vertical dimension, how can we reorient the desired image 90deg in order to fit the subject into the wider (longer) view dimension?
Funny, I've mentioned always using the print screen button - and finishing up in Paint. By some divine co-inky-dink, last night I received notice that Paint will shortly be moving to the Microsoft store, where it will be made available for free. I suppose it will no longer be bundled with Microsoft products. Time marches on.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
I've added a new setting to change the orientation of the atom. It is still vertical by default, but can be changed to horizontal in the settings dialog.
The electron images look good.
I prefer the first comparison between P2 and S2. I think it looks better with a common center rather than a common top, even though the latter shows the difference better. I'm still thinking about removing the difference by reducing the space taken up by the neutrons.
The electron images look good.
I prefer the first comparison between P2 and S2. I think it looks better with a common center rather than a common top, even though the latter shows the difference better. I'm still thinking about removing the difference by reducing the space taken up by the neutrons.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
That's pretty good, Airman. A few things to change:
null - no stack in first attach position
This applies to any position, not just the first. Most times it makes no difference, but it does with the Carousel level, as it can have 4 items.
Separation of particles or stacks without a bond
This actually separates atoms, not particles or stacks.
Similarly with flip and rotate, they only work on atoms.
Link between stacks - to create chains of alternating directions - NOT WORKING
Chains like that are not meant to work in isolation. You can only use chains in the Axial and Carousel levels (and the equivalent X, Y and Z). Since an atom requires a single stack for its core, you can not use chains alone.
Maybe you could add some higher level structures to show how the different parts of the expression fit together. For example, the top level entity is a multi-nuclei or normal atom. A multi-nuclei atom contains multiple atoms. Each atom contains a core and several levels. Each level can contain nuclear chains. Each nuclear chain can contain particle chains (which the default stacks are a version of). Each particle chain can contain particles. Somehow show the hierarchy of entities. Maybe in its own chart. Maybe it can fit on this one.
null - no stack in first attach position
This applies to any position, not just the first. Most times it makes no difference, but it does with the Carousel level, as it can have 4 items.
Separation of particles or stacks without a bond
This actually separates atoms, not particles or stacks.
Similarly with flip and rotate, they only work on atoms.
Link between stacks - to create chains of alternating directions - NOT WORKING
Chains like that are not meant to work in isolation. You can only use chains in the Axial and Carousel levels (and the equivalent X, Y and Z). Since an atom requires a single stack for its core, you can not use chains alone.
Maybe you could add some higher level structures to show how the different parts of the expression fit together. For example, the top level entity is a multi-nuclei or normal atom. A multi-nuclei atom contains multiple atoms. Each atom contains a core and several levels. Each level can contain nuclear chains. Each nuclear chain can contain particle chains (which the default stacks are a version of). Each particle chain can contain particles. Somehow show the hierarchy of entities. Maybe in its own chart. Maybe it can fit on this one.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Last edited by LongtimeAirman on Tue Jan 15, 2019 8:34 pm; edited 2 times in total (Reason for editing : Added P.S.)
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Nice, but I think we need to differentiate between atoms and nuclei. In this context, they are actually the same thing, but it will help to use the terms in different ways. The difference? An atom is an individual entity, it can be defined and rendered by an AML expression. A nucleus is an atom that is a part of a multi-nuclei atom. So, in essence, if an atom is being used in a multi-nuclei atom, then it is a nuclei. If it is being used on its own, then it is an atom.
Separation of atoms without a bond
should become: Separation of nuclei in a multi-nuclei atom
Similarly, the flip and rotate descriptions can use nuclei instead of atom.
I think it might be beneficial to create sections in order to see how certain operators only work in certain parts of an expression. This will probably satisfy what I mentioned at the end of my last post. Create a Particle Chain section, which contains both particles and particle chains, the : operator and multiplication groups. Then a Nuclear Chain section that contains default stacks (maybe put into Particle Chain section), the - operator and multiplication groups (yes, duplicate them). then create an Atom section that contains atoms and levels. Finally, create a Multi-nuclei Atom section for multi-nuclei atoms, ~, ! and ^ operators.
If you think it worth while, then create a Particle section first, then Particle Chain, etc.
Separation of atoms without a bond
should become: Separation of nuclei in a multi-nuclei atom
Similarly, the flip and rotate descriptions can use nuclei instead of atom.
I think it might be beneficial to create sections in order to see how certain operators only work in certain parts of an expression. This will probably satisfy what I mentioned at the end of my last post. Create a Particle Chain section, which contains both particles and particle chains, the : operator and multiplication groups. Then a Nuclear Chain section that contains default stacks (maybe put into Particle Chain section), the - operator and multiplication groups (yes, duplicate them). then create an Atom section that contains atoms and levels. Finally, create a Multi-nuclei Atom section for multi-nuclei atoms, ~, ! and ^ operators.
If you think it worth while, then create a Particle section first, then Particle Chain, etc.
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
.
Nevyn, I like your latest, opening AML - https://www.nevyns-lab.com/mathis/app/aml/aml.html with a quick "How To", as well as a link to the Specification. https://www.nevyns-lab.com/mathis/app/aml/spec.php. This is a great document. The 'reference sheet' seems a bit superfluous now; nevertheless, I'll try to expand it in accordance with the Specification, then you can decide to keep it or not.
One question - anomaly, sometimes the AML code results in off-center images, such as the 3 images in the the Specification, Multiplying within a Nuclear Chain section. I suppose it's just the view's y dimension calculation hasn't been completed sufficiently to accommodate the Nuclei Multiplication groups.
.
Nevyn, I like your latest, opening AML - https://www.nevyns-lab.com/mathis/app/aml/aml.html with a quick "How To", as well as a link to the Specification. https://www.nevyns-lab.com/mathis/app/aml/spec.php. This is a great document. The 'reference sheet' seems a bit superfluous now; nevertheless, I'll try to expand it in accordance with the Specification, then you can decide to keep it or not.
One question - anomaly, sometimes the AML code results in off-center images, such as the 3 images in the the Specification, Multiplying within a Nuclear Chain section. I suppose it's just the view's y dimension calculation hasn't been completed sufficiently to accommodate the Nuclei Multiplication groups.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
The offset images, I believe, are caused by using the arrow keys while editing the expression. Unfortunately, both the input text field and the controls for the camera hear the key events and respond to them. I have tried to fix it in the past, but no success. I shall try again. I was only thinking about that last night too.
As far as atom placement goes, all atoms are centered on their own axis. So even if you have uneven numbers of proton stacks on, say, the east and west, it will still be centered on the atom, not the size of it.
Your reference chart is still extremely valuable. The spec and the chart do different things. The spec is there for those that know nothing of the language and need to be shown how to build an expression. The chart is there for those that already know the language, but just need a quick reminder of the operators and structure. I might turn it into HTML though, rather than an image. That will make it more maintainable and flexible.
As far as atom placement goes, all atoms are centered on their own axis. So even if you have uneven numbers of proton stacks on, say, the east and west, it will still be centered on the atom, not the size of it.
Your reference chart is still extremely valuable. The spec and the chart do different things. The spec is there for those that know nothing of the language and need to be shown how to build an expression. The chart is there for those that already know the language, but just need a quick reminder of the operators and structure. I might turn it into HTML though, rather than an image. That will make it more maintainable and flexible.
Last edited by Nevyn on Wed Jan 16, 2019 5:00 pm; edited 1 time in total
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
Language Specification
I have created a specification for the language, although it reads more like a tutorial than a spec but I don't mind that. It is littered with images to show the structures as I present them.
If you look closely, you might notice that I have managed to generate nuclear chains without being a part of an atom. To do this, you can start the expression with either a - or a + character. These will both signal to the parser that it should treat the expression as a nuclear chain instead of an atom. It also sets a property on the chain that tells the visualizer that this is an axial or carousel chain. The difference is the orientation of the first stack (and all subsequent stacks).
To generate a carousel stack, use the - character: -S6-S4-S2
To generate an axial stack, use the + character: +S6-S4-S2
www.nevyns-lab.com/mathis/app/aml/spec.php
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
I added a couple of new sections to the spec about neutron and electron groups. I used your images, Airman, thanks for that. I also fixed a few errors and changed a few lines here or there, but nothing major. I think it is all looking good and am just about to launch it. Which means I need to think about some graphics for the front page, hmm, ...
 Re: Atomic Modeling Language
Re: Atomic Modeling Language
..., and there it goes, sailing off into the world after the official launch. What places will it see? Who will it meet? Only time will tell.
www.nevyns-lab.com/
www.nevyns-lab.com/
Page 1 of 2 • 1, 2 
 Similar topics
Similar topics» Modeling a Charge Particle
» Spinning Particle Language
» Molecular Bonding Language
» Atomic Model Editor
» Natural Language Processing (NLP) and newer algorithms
» Spinning Particle Language
» Molecular Bonding Language
» Atomic Model Editor
» Natural Language Processing (NLP) and newer algorithms
Page 1 of 2
Permissions in this forum:
You cannot reply to topics in this forum