Molecular Interactions (Noncovalent Interactions) - with RNA/DNA diagrams
Miles Mathis' Charge Field :: Miles Mathis Charge Field :: The Charge Field Effects on Humans/Animals
Page 1 of 1
 Molecular Interactions (Noncovalent Interactions) - with RNA/DNA diagrams
Molecular Interactions (Noncovalent Interactions) - with RNA/DNA diagrams
A pretty good paper.
Molecular Interactions (Noncovalent Interactions)
illustrated by biochemical systems
http://ww2.chemistry.gatech.edu/~lw26/structure/molecular_interactions/mol_int.html#B
Structure Tool:
MACROMOLECULAR STRUCTURE PROTEINS AND NUCLEIC ACIDS (uses a program called pymol)
http://ww2.chemistry.gatech.edu/~lw26/structure/index.html
Loren Williams wrote:
The development of this document has been supported by the NASA Astrobiology Institute, the National Science Foundation, and the School of Chemistry and Biochemistry at Georgia Tech, all of whom have supported my research laboratory and my public outreach efforts. Comments and suggestions for improvements are welcome and should be addressed to ldw@gatech.edu. The figures and text can be used with attribution for noncommercial purposes. I am hopeful that students, especially those who lack resources for textbooks, find this site to be useful.
Sincerely,
Loren Williams,
Professor
Georgia Tech
B. Short Range Repulsion.
| Table of Contents | Structure Tool | Williams Home |
Atoms take space. If two atoms are forced together, they push back. As two atoms approach each other, at some distance the occupied orbitals begin to overlap, causing electrostatic repulsion between the electrons. This repulsive force between atoms acts over a very short range, but goes up sharply when that range is violated.
The repulsion goes up as 1/R12, where R is the distance between the atoms. The large exponent means small changes in R cause large changes in the repulsion. Short range repulsion is important only when atoms are in very close proximity, but at close range it is very important. Because this repulsive term rises so sharply as distance decreases it is often useful to pretend that atom are hard spheres, like very small pool balls, with hard surfaces (called van der Waals surfaces) and well-defined radii (r, called van der Waals radii).
As two atoms approach each other their van der Waals surfaces make contact when the distance between them equals the sum of their van der Waals radii. At this distance the repulsive energy skyrockets. The smallest distance between two non-bonded atoms is the sum of the van der Waals radii of the two atoms. A sulfur atom and a carbon atom can come no closer together than:
rS + rC = 1.8 + 1.7 = 3.5 Å.
Of course we are assuming here that bonds do not form. When two atoms form a bond, they come very close together and their der Waals radii and surfaces are violated.
Short range repulsion is important to you. It prevents your hands from passing through each other when you clap, and prevents atoms from collapsing into tightly packed states of enormous density of 1014 g/ml, which is the density of condensed atomic nuclei. Very high gravity, as on neutron stars, overwhelm short range repulsion and cause atoms to collapse.
Here in earth, with our modest gravity, the van der Waals radius of carbon (rC) is evident from the spacing between the layers in graphite. The distance between atoms in different layers of graphite is never less than twice the van der Waals radius of carbon (2 x rC = 2 x 1.7 = 3.4 Å). The atoms within a graphite layer are covalently linked (bonded), which causes interpenetration of van der Waals surfaces. Carbon atoms within a layer are separated by 1.42 Å, which is much less than twice the van der Waals radius of carbon. As explained in other sections of this document vdw surfaces are also violated when molecules form hydrogen bonds.The coordinates of graphite are here [coordinates].
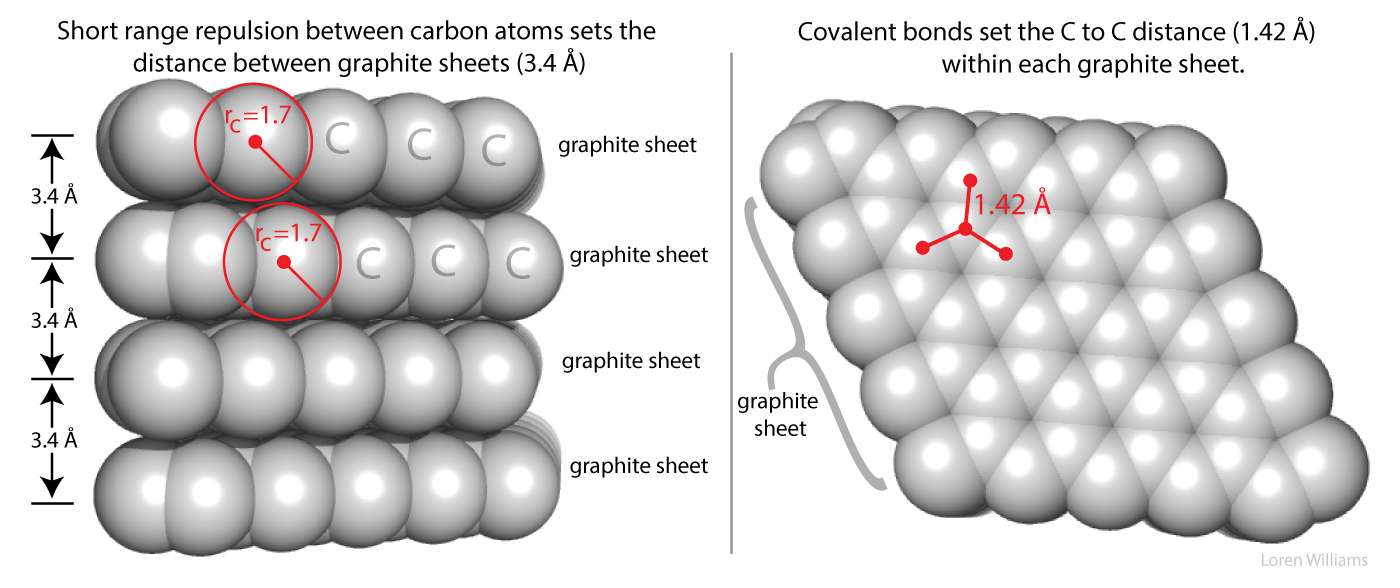
Figure 2 shows how short range repulsion sets the distance of 3.4 Å between sheets in graphite. If two non-bonded atoms are separated by a distance of less than the sum of their VDW radii, short range repulsion forces them apart. This image can be reused with attribution for noncommercial purposes.
In B-DNA, the helical rise per base pair of 3.4 Å is determined by short range repulsion between the stacked bases. Bases stack on each other with spacing of 3.4 Å.
How do you sense short range repulsion? Try compressing a liquid.
C. Electrostatic Interactions.
Electrostatic interactions are the primary stabilizing interaction between phosphate oxygens of RNA (formal charge -1) and magnesium ions (formal charge +2), as shown in the figure below. There are many magnesium ions associated with RNA and DNA molecules in vivo . As explained later in this document, electrostatic interactions are highly attenuated (dampened) by water. In protein folding, RNA folding and DNA annealing, electrostatic interactions are dependent on salt concentration and pH.
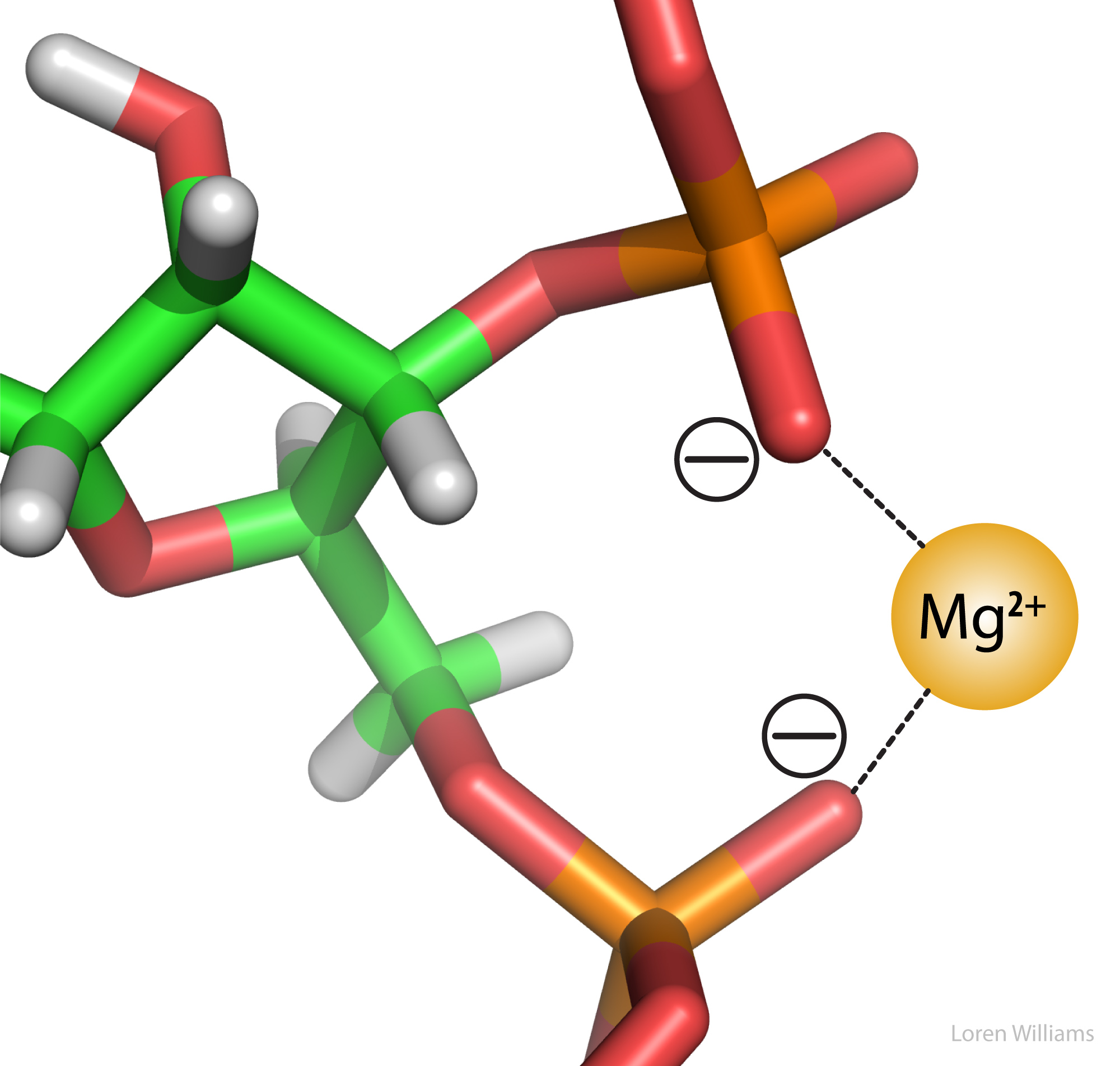
 Re: Molecular Interactions (Noncovalent Interactions) - with RNA/DNA diagrams
Re: Molecular Interactions (Noncovalent Interactions) - with RNA/DNA diagrams
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4254145/
(much more at link....)
Published online 2014 Jul 5. doi: 10.1016/j.gde.2014.05.005
PMCID: PMC4254145
NIHMSID: NIHMS611650
Sirtuins, Metabolism, and DNA repair
[url=https://www.ncbi.nlm.nih.gov/pubmed/?term=Choi JE[auth]]Jee-Eun Choi[/url] and [url=https://www.ncbi.nlm.nih.gov/pubmed/?term=Mostoslavsky R[auth]]Raul Mostoslavsky[/url]*
Author information ► Copyright and License information ►
The publisher's final edited version of this article is available at Curr Opin Genet Dev
See other articles in PMC that cite the published article.
Cells evolve to actively coordinate nutrient availability with cellular activity in order to maintain metabolic homeostasis. In addition, active pathways to repair DNA damage are crucial to avoid deleterious genomic instability. In recent years, it has become increasingly clear that availability of intermediate metabolites may play an important role in DNA repair, suggesting that these two seemingly distant cellular activities may be highly coordinated. The sirtuin family of proteins now described as deacylases (they can also remove acyl groups other than acetyl moieties), it appears to have evolved to control both metabolism and DNA repair. In this review, we discuss recent advances that lay the foundation to understanding the role of sirtuins in these two biological processes, and the potential crosstalk to coordinate them.
Sirtuins are members of a family of evolutionarily conserved enzymes with NAD+-dependent deacylase activity. Since the discovery of Sir2 (silencing information regulator 2) in the budding yeast Saccharomyces cerevisiae as a transcriptional silencer of the mating-type loci more than 20 years ago [1], many studies have demonstrated diverse biological roles for sirtuins, such as in genome stability, cellular metabolism, and lifespan regulation [2,3]. Mammalian sirtuins have seven isoforms (SIRT1–7), each one with unique subcellular localization and distinct functions [4]. SIRT1 and SIRT2 can be found in both nucleus and cytoplasm, SIRT6 and SIRT7 are almost exclusively nuclear and SIRT3, SIRT4, and SIRT5 are located in the mitochondria [5]. Studies on sirtuin biology have shown great progress in the past two decades, emphasizing the critical importance of these enzymes in human biology and disease.
Due to their NAD+ dependency, it had been speculated that sirtuins play a crucial role in modulating energy metabolism. Indeed, sirtuins are broadly recognized as critical regulators of multiple metabolic pathways, including glucose, glutamine, and lipid metabolism [6]. For cells to thrive, energy and metabolic demands have to be carefully coordinated with nutrients availability. As sensors of energy and redox status in cells, these protein deacylases can directly modulate activity of key metabolic enzymes -by posttranslational modifications- as well as regulate transcription of metabolic genes. In addition, several sirtuins play additional roles in metabolic homeostasis. For instance, both SIRT1 and SIRT2 control autophagy responses under various nutrient stress conditions, as modulators of FOXO signaling pathway [7]. Autophagy will be covered in detail in an accompanying article in this issue.
Nuclear sirtuins have also evolved as regulators of genome integrity. Our cells experience ~ 1×104−1×105 DNA lesions per day [8], hence they have developed repair machineries to avoid detrimental outcomes from oxidative and genotoxic stress. In the past decade, the roles of sirtuins in maintaining genomic stability have been described, as regulators of DNA repair pathways [9], chromatin structure [10], and telomere maintenance [11,12].
Based on the fact that sirtuins possess dual roles in metabolism and DNA repair, sirtuins can serve as nodal points in regulating both processes. Intriguingly, new studies have started to appreciate that DNA damage can directly trigger adaptive metabolic responses [13,14], indicating that these two seemingly separate biological entities may function in a highly coordinated fashion. In this review, we will focus on recent progress in understanding the roles of sirtuins in both metabolism and DNA repair, and the possible crosstalk between these two phenomena.
Since glucose is a primary nutrient for cell survival and proliferation, systemic glucose levels should be tightly regulated throughout tissues. Crucial organs such as liver, muscle, and pancreas are main modulators of glucose homeostasis. At the cellular level, once glucose enters a cell, it is converted into pyruvate in the cytoplasm through glycolysis in a multi-enzyme, strictly regulated process. In most cells, pyruvate will then enter the TCA cycle to generate energy through oxidative phosphorylation (OXPHOS) in a highly efficient process (34–36 mols of ATP per mol of glucose). However, in specific cases, pyruvate will be diverted in the cytoplasm to produce lactate, a less efficient way to produce ATP, but a critical adaptive mechanism in cells where OXPHOS is impeded (hypoxia, for instance) or to produce intermediate metabolites for biomass in highly proliferating cells.
Extensive studies have previously shown that SIRT1 can modulate both gluconeogenesis and glycolysis by regulating important metabolic factors, including PGC1α and FOXO [15]. More recently, intracellular levels of NAD+ has been shown to regulate SIRT1 deacetylase activity, affecting high fat diet (HFD)-induced obesity and aging, as discussed below [reviewed in 16].
SIRT3 is a major mitochondrial protein deacetylase [17], regulating multiple metabolic proteins such as the TCA cycle protein isocitrate dehydrogenease 2 (IDH2) [18] and key proteins in the electron transfer chain (ETC) [19–21]. In skeletal muscle, SIRT3 plays an important role in regulating metabolic adaptive responses. Decreased levels of SIRT3 cause increasing oxidative stress and insulin resistance [22] and recent studies showed that active deacetylation of pyruvate dehydrogenase (PDH) E1α by SIRT3 provides metabolic flexibility under nutrient stress conditions [23]. Wang and his colleagues discovered that SIRT3 can deacetylate FOXO3a, in turn enhancing FOXO3a activity and increased expression of its targets, including antioxidant genes. In this way, SIRT3 protects mitochondria from oxidative stress [24]. Since SIRT3 actively modulate carbohydrate metabolism and ROS production, the role of SIRT3 in cancer metabolism has been highlighted [25]. Gius et al. first described that SIRT3 acts as a tumor suppressor by maintaining intact mitochondria in breast cancer [26]. Later, two studies provided mechanistic proof that HIF-1α (hypoxia inducible factor-1α) stabilization following mitochondrial ROS generation is critical to sustain cancer-prone metabolic reprogramming in SIRT3-deleted tumors [27,28].
SIRT4 is mostly known for its role in glutamine metabolism. In proliferating cells, glutamine is the main source to replenish the TCA cycle as a source of α-ketoglutarate (α-KG) [29]. Two different groups recently reported new roles for SIRT4 in glutamine metabolism. Jeong et al. described that SIRT4 inhibits glutamine entry to the TCA cycle under genotoxic stress, preventing dysregulated proliferation and genomic instability [14]. Although SIRT4 appears to work by inhibiting GDH activity, how SIRT4 does so mechanistically remains to be fully understood. Notably, Csibi et al. found that the mTORC1-CREB2 axis can regulate SIRT4 transcription under various nutrient stress conditions, thereby affecting glutamine anaplerosis into the TCA cycle and cell proliferation [30], further confirming an important role for this sirtuin in glutamine metabolism.
SIRT5 has recently been defined as a lysine demalonylase and desuccinylase [31]. The global analysis of lysine succinylation (“succinylome”) in the context of SIRT5 demonstrated that this posttranslational modification has a regulatory effect on glucose metabolism by modulating the activities of PDH, SDH and mitochondrial respiration in mouse liver and MEFs [32]. The pioneering work of the Lin laboratory provided the first proof that sirtuins can work by removing non-acetyl acyl groups, defining sirtuins as “protein deacylases” and opening a whole new field in enzymology and biochemistry.
(much more at link....)
Published online 2014 Jul 5. doi: 10.1016/j.gde.2014.05.005
PMCID: PMC4254145
NIHMSID: NIHMS611650
Sirtuins, Metabolism, and DNA repair
[url=https://www.ncbi.nlm.nih.gov/pubmed/?term=Choi JE[auth]]Jee-Eun Choi[/url] and [url=https://www.ncbi.nlm.nih.gov/pubmed/?term=Mostoslavsky R[auth]]Raul Mostoslavsky[/url]*
Author information ► Copyright and License information ►
The publisher's final edited version of this article is available at Curr Opin Genet Dev
See other articles in PMC that cite the published article.
Abstract
Cells evolve to actively coordinate nutrient availability with cellular activity in order to maintain metabolic homeostasis. In addition, active pathways to repair DNA damage are crucial to avoid deleterious genomic instability. In recent years, it has become increasingly clear that availability of intermediate metabolites may play an important role in DNA repair, suggesting that these two seemingly distant cellular activities may be highly coordinated. The sirtuin family of proteins now described as deacylases (they can also remove acyl groups other than acetyl moieties), it appears to have evolved to control both metabolism and DNA repair. In this review, we discuss recent advances that lay the foundation to understanding the role of sirtuins in these two biological processes, and the potential crosstalk to coordinate them.
Introduction
Sirtuins are members of a family of evolutionarily conserved enzymes with NAD+-dependent deacylase activity. Since the discovery of Sir2 (silencing information regulator 2) in the budding yeast Saccharomyces cerevisiae as a transcriptional silencer of the mating-type loci more than 20 years ago [1], many studies have demonstrated diverse biological roles for sirtuins, such as in genome stability, cellular metabolism, and lifespan regulation [2,3]. Mammalian sirtuins have seven isoforms (SIRT1–7), each one with unique subcellular localization and distinct functions [4]. SIRT1 and SIRT2 can be found in both nucleus and cytoplasm, SIRT6 and SIRT7 are almost exclusively nuclear and SIRT3, SIRT4, and SIRT5 are located in the mitochondria [5]. Studies on sirtuin biology have shown great progress in the past two decades, emphasizing the critical importance of these enzymes in human biology and disease.
Due to their NAD+ dependency, it had been speculated that sirtuins play a crucial role in modulating energy metabolism. Indeed, sirtuins are broadly recognized as critical regulators of multiple metabolic pathways, including glucose, glutamine, and lipid metabolism [6]. For cells to thrive, energy and metabolic demands have to be carefully coordinated with nutrients availability. As sensors of energy and redox status in cells, these protein deacylases can directly modulate activity of key metabolic enzymes -by posttranslational modifications- as well as regulate transcription of metabolic genes. In addition, several sirtuins play additional roles in metabolic homeostasis. For instance, both SIRT1 and SIRT2 control autophagy responses under various nutrient stress conditions, as modulators of FOXO signaling pathway [7]. Autophagy will be covered in detail in an accompanying article in this issue.
Nuclear sirtuins have also evolved as regulators of genome integrity. Our cells experience ~ 1×104−1×105 DNA lesions per day [8], hence they have developed repair machineries to avoid detrimental outcomes from oxidative and genotoxic stress. In the past decade, the roles of sirtuins in maintaining genomic stability have been described, as regulators of DNA repair pathways [9], chromatin structure [10], and telomere maintenance [11,12].
Based on the fact that sirtuins possess dual roles in metabolism and DNA repair, sirtuins can serve as nodal points in regulating both processes. Intriguingly, new studies have started to appreciate that DNA damage can directly trigger adaptive metabolic responses [13,14], indicating that these two seemingly separate biological entities may function in a highly coordinated fashion. In this review, we will focus on recent progress in understanding the roles of sirtuins in both metabolism and DNA repair, and the possible crosstalk between these two phenomena.
Sirtuins in metabolism
Glucose and glutamine metabolism
Since glucose is a primary nutrient for cell survival and proliferation, systemic glucose levels should be tightly regulated throughout tissues. Crucial organs such as liver, muscle, and pancreas are main modulators of glucose homeostasis. At the cellular level, once glucose enters a cell, it is converted into pyruvate in the cytoplasm through glycolysis in a multi-enzyme, strictly regulated process. In most cells, pyruvate will then enter the TCA cycle to generate energy through oxidative phosphorylation (OXPHOS) in a highly efficient process (34–36 mols of ATP per mol of glucose). However, in specific cases, pyruvate will be diverted in the cytoplasm to produce lactate, a less efficient way to produce ATP, but a critical adaptive mechanism in cells where OXPHOS is impeded (hypoxia, for instance) or to produce intermediate metabolites for biomass in highly proliferating cells.
Extensive studies have previously shown that SIRT1 can modulate both gluconeogenesis and glycolysis by regulating important metabolic factors, including PGC1α and FOXO [15]. More recently, intracellular levels of NAD+ has been shown to regulate SIRT1 deacetylase activity, affecting high fat diet (HFD)-induced obesity and aging, as discussed below [reviewed in 16].
SIRT3 is a major mitochondrial protein deacetylase [17], regulating multiple metabolic proteins such as the TCA cycle protein isocitrate dehydrogenease 2 (IDH2) [18] and key proteins in the electron transfer chain (ETC) [19–21]. In skeletal muscle, SIRT3 plays an important role in regulating metabolic adaptive responses. Decreased levels of SIRT3 cause increasing oxidative stress and insulin resistance [22] and recent studies showed that active deacetylation of pyruvate dehydrogenase (PDH) E1α by SIRT3 provides metabolic flexibility under nutrient stress conditions [23]. Wang and his colleagues discovered that SIRT3 can deacetylate FOXO3a, in turn enhancing FOXO3a activity and increased expression of its targets, including antioxidant genes. In this way, SIRT3 protects mitochondria from oxidative stress [24]. Since SIRT3 actively modulate carbohydrate metabolism and ROS production, the role of SIRT3 in cancer metabolism has been highlighted [25]. Gius et al. first described that SIRT3 acts as a tumor suppressor by maintaining intact mitochondria in breast cancer [26]. Later, two studies provided mechanistic proof that HIF-1α (hypoxia inducible factor-1α) stabilization following mitochondrial ROS generation is critical to sustain cancer-prone metabolic reprogramming in SIRT3-deleted tumors [27,28].
SIRT4 is mostly known for its role in glutamine metabolism. In proliferating cells, glutamine is the main source to replenish the TCA cycle as a source of α-ketoglutarate (α-KG) [29]. Two different groups recently reported new roles for SIRT4 in glutamine metabolism. Jeong et al. described that SIRT4 inhibits glutamine entry to the TCA cycle under genotoxic stress, preventing dysregulated proliferation and genomic instability [14]. Although SIRT4 appears to work by inhibiting GDH activity, how SIRT4 does so mechanistically remains to be fully understood. Notably, Csibi et al. found that the mTORC1-CREB2 axis can regulate SIRT4 transcription under various nutrient stress conditions, thereby affecting glutamine anaplerosis into the TCA cycle and cell proliferation [30], further confirming an important role for this sirtuin in glutamine metabolism.
SIRT5 has recently been defined as a lysine demalonylase and desuccinylase [31]. The global analysis of lysine succinylation (“succinylome”) in the context of SIRT5 demonstrated that this posttranslational modification has a regulatory effect on glucose metabolism by modulating the activities of PDH, SDH and mitochondrial respiration in mouse liver and MEFs [32]. The pioneering work of the Lin laboratory provided the first proof that sirtuins can work by removing non-acetyl acyl groups, defining sirtuins as “protein deacylases” and opening a whole new field in enzymology and biochemistry.
 Similar topics
Similar topics» Collaborate to Promote Mathis' Ideas Better
» Molecular Structure of Acids
» What Will the Future of Molecular Manufacturing Really Be Like?
» Molecular Bonding Language
» Miles Periodic Table with Standard Periodic Table reference
» Molecular Structure of Acids
» What Will the Future of Molecular Manufacturing Really Be Like?
» Molecular Bonding Language
» Miles Periodic Table with Standard Periodic Table reference
Miles Mathis' Charge Field :: Miles Mathis Charge Field :: The Charge Field Effects on Humans/Animals
Page 1 of 1
Permissions in this forum:
You cannot reply to topics in this forum