Molecular Bonding Language
4 posters
Page 2 of 2
Page 2 of 2 •  1, 2
1, 2
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Jesus, this is some amazing and very rapid progress! Pretty incredible stuff here, Nevyn. I'm digging in a bit and it's a lot of fun, as well as looking really cool. And a very interesting way to visualize molecular structures!
Jared Magneson- Posts : 525
Join date : 2016-10-11
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Thanks guys. When I'm on a roll, it just feels like the app falls together. New features just seem to fit right in. By creating a language, parser and renderer, it has allowed me to separate different aspects cleanly. Before this, I had always thought in terms of a builder. Allowing the user to add pieces and place them but that would take a lot of work, both for me to create the app and for the user to do anything with it. A language is extremely easy to use and change and play with.
If you go over old posts here, you'll see that Cr6 has been wanting something like this for years but I just wasn't seeing it. It didn't resonate with me until the recent posts on super-conductors and I'm so glad it finally did. I guess this was just the right time for it to come together. I had some pieces and just needed to plug them into a new engine.
I've been keeping AtomicViewer up-to-date with the changes I make for MBL and have even spent some time going over some nuclear models again, which is long overdue. Seriously, they need a lot of work. Feel free to criticize any of them. If you are building molecules and find an atom isn't working for you, then it may mean that the atom is incorrect. Especially with the large atoms. A good way to test the nuclear model of an atom is to see how it works in known molecules. MBL makes that so much easier!
How awesome does that carbon nano-tube look above? Go and create one now! Play with it. Spin it around. Move through it. And take pictures. I wanna see em. Bring out your inner-photographer and capture the beauty of these wonderful structures.
If you go over old posts here, you'll see that Cr6 has been wanting something like this for years but I just wasn't seeing it. It didn't resonate with me until the recent posts on super-conductors and I'm so glad it finally did. I guess this was just the right time for it to come together. I had some pieces and just needed to plug them into a new engine.
I've been keeping AtomicViewer up-to-date with the changes I make for MBL and have even spent some time going over some nuclear models again, which is long overdue. Seriously, they need a lot of work. Feel free to criticize any of them. If you are building molecules and find an atom isn't working for you, then it may mean that the atom is incorrect. Especially with the large atoms. A good way to test the nuclear model of an atom is to see how it works in known molecules. MBL makes that so much easier!
How awesome does that carbon nano-tube look above? Go and create one now! Play with it. Spin it around. Move through it. And take pictures. I wanna see em. Bring out your inner-photographer and capture the beauty of these wonderful structures.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
It actually works really smoothly on my simple A8 (quad-core) TV-PC, with just a GTX-750Ti for the graphics. It's about a $50 graphics chip now, the only one I have small enough for this home theater case, but I'm gonna try it on my 'Dozer with a GTX-980 when I get some time, and push it much harder!
I just don't know enough about molecular structures to come up with anything too cool or realistic, so I'm just lobbing atoms out there to see how they look.
Here's my stab at some Germanium-Silicon nonsense, just based on your carbon nanotube description:

MBL: tube 6 n:(C{Ge,C,Si}C)6
And in Line form:

I just don't know enough about molecular structures to come up with anything too cool or realistic, so I'm just lobbing atoms out there to see how they look.
Here's my stab at some Germanium-Silicon nonsense, just based on your carbon nanotube description:

MBL: tube 6 n:(C{Ge,C,Si}C)6
And in Line form:

Jared Magneson- Posts : 525
Join date : 2016-10-11
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
.

Nevyn. Congratulations, this is great. Honest to gosh, I got playground flashbacks. Like I was playing on the large metal climbing structure at the playground when I was six.
Back to the present, I make word doc copies of all new posts before I read them. When I highlighted, copied and switched to the browser in order to paste the MBL tag - tube 6 n:(C{C,C}C)10 , I bombed Word and my system. I realized the tag was a link that wouldn’t allow the word doc to reload properly. No problem, the second time I replaced each C with Si to create an image as you requested. I selected the “middle” choice for image sizes; the downloaded image is 3.4M - too large to share - so I screen dumped it at reduced magnification to a more manageable 850K png file.
I didn’t look at MBL too critically. For example, Language Specification needs examples and images. I’ll stop there. I don’t feel qualified to make a critical review just yet; that is, if you want one.
How about you Cr6? You’ve got a good handle on it. Care to make any critical remarks or suggestions?
.

Nevyn. Congratulations, this is great. Honest to gosh, I got playground flashbacks. Like I was playing on the large metal climbing structure at the playground when I was six.
Back to the present, I make word doc copies of all new posts before I read them. When I highlighted, copied and switched to the browser in order to paste the MBL tag - tube 6 n:(C{C,C}C)10 , I bombed Word and my system. I realized the tag was a link that wouldn’t allow the word doc to reload properly. No problem, the second time I replaced each C with Si to create an image as you requested. I selected the “middle” choice for image sizes; the downloaded image is 3.4M - too large to share - so I screen dumped it at reduced magnification to a more manageable 850K png file.
I didn’t look at MBL too critically. For example, Language Specification needs examples and images. I’ll stop there. I don’t feel qualified to make a critical review just yet; that is, if you want one.
How about you Cr6? You’ve got a good handle on it. Care to make any critical remarks or suggestions?
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
That's cool, Airman, just create stuff that looks good. No need to worry about being correct at this point. There's plenty of time for us to work on actual molecules.
Your imagery of playground equipment has given me a new idea for a Chain Command called 'mirror' that will take a defined chain and create a mirrored version next to it. You made me think about Monkey Bars (do you call them that in the states?) where you swing from bar to bar (no, not from pub to pub, that's the adult version ).
).
I use 720 for my screenshots. The others are too large but great if you want to study them as you can zoom in really far without losing quality. Even the 720 images are a bit too big (pixel wise) for a forum post which seem to prefer images about 800 pixels wide (the 720 refers to the Y axis, not the X). I might add a 480 res.
Reviews and criticism are always welcome. Images are indeed coming for the specification.
Your imagery of playground equipment has given me a new idea for a Chain Command called 'mirror' that will take a defined chain and create a mirrored version next to it. You made me think about Monkey Bars (do you call them that in the states?) where you swing from bar to bar (no, not from pub to pub, that's the adult version
I use 720 for my screenshots. The others are too large but great if you want to study them as you can zoom in really far without losing quality. Even the 720 images are a bit too big (pixel wise) for a forum post which seem to prefer images about 800 pixels wide (the 720 refers to the Y axis, not the X). I might add a 480 res.
Reviews and criticism are always welcome. Images are indeed coming for the specification.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
I think it's great that you included a render-to-file option! The 1080 size is fine resolution-wise, it's just an uncompressed .png so it's a much bigger file than it could be. I just convert to .jpg locally and upload them to Imgur or whatever anyway, all my tools are Shell extensions in Windows so it's pretty seamless, but most people won't have them tools handy. Damnit, I'm starting to write with a Southern "accent" even though I've never had one! Too much Twilight Zone lately.
I don't know how easy it would be for you to outpout straight to .jpg, but if that's an option most people might prefer it. Or compressed .png. I get why you chose the one you did though; it's a much cleaner file, with less artifacting.
I don't know how easy it would be for you to outpout straight to .jpg, but if that's an option most people might prefer it. Or compressed .png. I get why you chose the one you did though; it's a much cleaner file, with less artifacting.
Jared Magneson- Posts : 525
Join date : 2016-10-11
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
.
I usually use screen dumps; however I very much appreciate having the rendering download options - much better quality, bigger than my screen. I usually go with png's because this site doesn't allow me to post large jpg files.
I enjoyed the Outer Limits episode.
Excuse the blast from the past. They don't exactly fit well with your beautiful molecule images. Both are taken from When Playgrounds Were Deadly https://flashbak.com/when-playgrounds-were-deadly-35924/
We had something similar to 6 of these moon climbers side by side, forming a half cylinder, that’s what we called “the monkey bars”.

We also had what’s pictured below, pretty much a standard playground appliance in NYC. If I recall correctly, the surface the was some sort of tar.
 .
.
I usually use screen dumps; however I very much appreciate having the rendering download options - much better quality, bigger than my screen. I usually go with png's because this site doesn't allow me to post large jpg files.
I enjoyed the Outer Limits episode.
Excuse the blast from the past. They don't exactly fit well with your beautiful molecule images. Both are taken from When Playgrounds Were Deadly https://flashbak.com/when-playgrounds-were-deadly-35924/
We had something similar to 6 of these moon climbers side by side, forming a half cylinder, that’s what we called “the monkey bars”.

We also had what’s pictured below, pretty much a standard playground appliance in NYC. If I recall correctly, the surface the was some sort of tar.

LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
I have added the ability to choose between PNG and JPEG. If you use JPEG then you can also set the quality which will make the files a lot smaller/larger. The default is a good trade-off even though it looks like it is too far to the right. You can also set the name of the file, the extension is set based on the selected format.

Note: That is not actually the default value in the above image. That is full quality and the default is 2 steps down from that.

Note: That is not actually the default value in the above image. That is full quality and the default is 2 steps down from that.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Nice and so very cool Nevyn! It is really fun to use! 
The only thing I could ask for at this point would be an ability (radio button) to show an exploded version of the MBL language when a Chem notation was typed in... something like:
CdNbBa9Cu10O20 --> Cd-Nb-(Ba)9-(Cu)10-(O)20 or all exploded into a long "....-O-O" type line of single atoms? Not entirely necessary but it might allow more direct editing in the MBL language?
It looks AWESOME and like LTAM has suggested previously.... I'm on the Playground!
I'm on the Playground!
The only thing I could ask for at this point would be an ability (radio button) to show an exploded version of the MBL language when a Chem notation was typed in... something like:
CdNbBa9Cu10O20 --> Cd-Nb-(Ba)9-(Cu)10-(O)20 or all exploded into a long "....-O-O" type line of single atoms? Not entirely necessary but it might allow more direct editing in the MBL language?
It looks AWESOME and like LTAM has suggested previously....
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Yeah, I've been wondering if you needed that kind of conversion or not. I allowed chem notation in the language and then thought that you might have wanted the converted expression rather than being able to get it into the MBL parser and renderer.
I'll come up with something.
I'll come up with something.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
You sure work fast! Just saw the new output options in practice, and was on my way here to chime in again.
I was reading this article on Solar energy breakthroughs:
https://www.sciencedaily.com/releases/2018/05/180524141611.htm
And they mentioned:
So I thought I'd try that structure out in MBL. I'm just guessing at how it would fit together here, but it sure is cool to visualize it so rapidly:

MBL:
Pb-Ti[O,O,O,O]Ti-F-Pb
I'm sure my assembly is WAY off, but it looks cool and is definitely an amazing study tool!
I was reading this article on Solar energy breakthroughs:
https://www.sciencedaily.com/releases/2018/05/180524141611.htm
And they mentioned:
Now, such efforts have taken a surprising turn, with the discovery of a new photocatalytic material called a pyrochlore[2] oxyfluoride (Pb2Ti2O5.4F1.2).
So I thought I'd try that structure out in MBL. I'm just guessing at how it would fit together here, but it sure is cool to visualize it so rapidly:

MBL:
Pb-Ti[O,O,O,O]Ti-F-Pb
I'm sure my assembly is WAY off, but it looks cool and is definitely an amazing study tool!
Jared Magneson- Posts : 525
Join date : 2016-10-11
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Language Update
Nuclear Sheets
I have added a new Chain Command that will create a sheet from a basic chain without using a multiplication group. This is more convenient when trying to find something that works but the multiplication version is better for converting to and from a tube. The definitions remain the same and only the chain command changes between line and tube but a sheet requires restructuring the expression.
sheet breadth height direction:chain
MBL: sheet:C{C,C}C
URL: https://www.nevyns-lab.com/mathis/app/mbl/mbl.html?mbl=sheet:C{C,C}C&align=X&atom=nucleus
MBL: sheet 5:C{C,C}C
MBL: sheet 5 8:C{C,C}C
MBL: sheet 5 8 e:C{C,C}C
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
UI Update
Labels
You can now specify a label to be rendered onto the image when you use the capture functionality. There is a button to copy the MBL into the label field and some positioning controls. The text is rendered at a consistent size even when using different resolutions. However, it will not change the size of the text even if it won't fit on the image. It will be drawn as a single line and will not split.


 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Chemical Formula Conversion
I have added a new button to convert a chemical formula to an MBL expression. It will only convert it if it does not contain any MBL delimiters, except for ( and ) which apply to both formats.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Nice!!! Thanks a bunch Nevyn...this makes it a whole lot easier to play with!
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Language Update
Chain Arrays
I have introduced a new concept into MBL to support declaring an array of chains. It is just a list of individual chains that can then be interpreted into something else by a Chain Command. To define a chain array, you use the '<' and '>' characters to surround a comma separated list of chains.
Example: <C,O-O,Fe[H,H]>
That would create an array with 3 items: C, O-O and Fe[H,H]. It would also generate an error if you tried to use it like that directly. We need a Chain Command that understands how to use an array.
Fans
You can use 3 new Chain Commands that require a Chain Array as input. Two of them are needed, the other not so much but it was easy once I had the others working.
Fan In
The fanIn command can be used to make all items in the list, except the last one, arrange themselves into a fan focused onto the last item in the array.
MBL: fanIn:<Cl,Cl,Na-Cl>
URL: https://www.nevyns-lab.com/mathis/app/mbl/mbl.html?mbl=fanIn:%3CCl,Cl,Na-Cl%3E&align=X&atom=nucleus

You can have as many items as you want, but they will get squashed up together. They are all evenly spaced apart with an angle equal to 180 divided by the number of items in the fan plus 1. That stops the outside chains from being placed vertically and will always have some angle away from the last chain.
Fan Out
Conversely, you can have the chains fan out from the first item in the array by using the fanOut command.
MBL: fanOut:<Na-Cl,!Cl,!Cl>
URL: https://www.nevyns-lab.com/mathis/app/mbl/mbl.html?mbl=fanOut:%3CNa-Cl,!Cl,!Cl%3E&align=X&atom=nucleus

You can even mix the 2 together like this:
MBL: fanIn:<Cl,Cl,fanOut:<Na-Cl,!Cl,!Cl>>
URL: https://www.nevyns-lab.com/mathis/app/mbl/mbl.html?mbl=fanIn:%3CCl,Cl,fanOut:%3CNa-Cl,!Cl,!Cl%3E%3E&align=X&atom=nucleus

Fan
While not useful for molecules, you can create pretty pictures using the fan command. This will just take all items in the array and arrange them in a circle.
MBL: fan:<(C{H,H}C,)4,C{H,H}C>
URL: https://www.nevyns-lab.com/mathis/app/mbl/mbl.html?mbl=fan:%3C(C{H,H}C,)4,C{H,H}C%3E&align=X&atom=nucleus
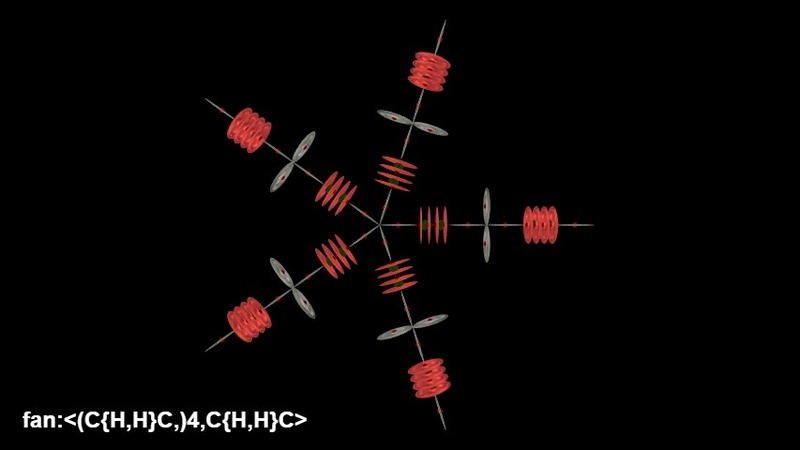
Uses for Fans
I needed to include these commands to support some of the structures that Miles has shown, such as NaCl3. It also means that we can model molecules like CH4 (Methane), but to do that you will need to break the Carbon atom up a bit so that the Hydrogens go into the south pole correctly. In order to do that, I will need to supply some broken atoms. For example, we need a Carbon atom that does not have its south Proton, as that will need to be specified as an extra H atom. I don't like having to break the atoms up like that, as it stops other possible functionality like extracting the chemical formula from the chain. However, if you just want it to look right, then it will be fine.
You can create some funky molecules like this:
MBL: fanIn:<fanIn:<O,O,Mg>,fanIn:<O,O,Mg>,fanOut:<Ba[fanOut:<Mg,O,O>,fanOut:<Mg,O,O>,fanOut:<Mg,O,O>,fanOut:<Mg,O,O>],fanOut:<Mg,O,O>,fanOut:<Mg,O,O>>>
URL: https://www.nevyns-lab.com/mathis/app/mbl/mbl.html?mbl=fanIn:%3CfanIn:%3CO,O,Mg%3E,fanIn:%3CO,O,Mg%3E,fanOut:%3CBa[fanOut:%3CMg,O,O%3E,fanOut:%3CMg,O,O%3E,fanOut:%3CMg,O,O%3E,fanOut:%3CMg,O,O%3E],fanOut:%3CMg,O,O%3E,fanOut:%3CMg,O,O%3E%3E%3E&align=X&atom=nucleus

Last edited by Nevyn on Sat Jun 16, 2018 7:40 pm; edited 2 times in total (Reason for editing : Chaned CH3 to CH4)
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Actually, the fan command is useful. I have used it to model Nitro-Glycerin years ago, which I modeled as 3 long arms attached at the center, as-in a Y configuration.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Forgot to mention that the fan* commands support some parameters.
fan* direction angle:chain array
where:
direction is north, south, east or west, or just the first letter, and dictates which way the atoms are placed and which direction they curve around. The default is east.
angle is the number of degrees to rotate around the chain axis, which effectively rotates the fan around its connection point to the focal chain. It is actually rotating the entire chain, not just the fan. Not sure if I will change that or not at this stage. Let me know if you think I should just rotate the fan and not the focal chain as well.
Example: fanIn n 45:<C,C,C,O>
fan* direction angle:chain array
where:
direction is north, south, east or west, or just the first letter, and dictates which way the atoms are placed and which direction they curve around. The default is east.
angle is the number of degrees to rotate around the chain axis, which effectively rotates the fan around its connection point to the focal chain. It is actually rotating the entire chain, not just the fan. Not sure if I will change that or not at this stage. Let me know if you think I should just rotate the fan and not the focal chain as well.
Example: fanIn n 45:<C,C,C,O>
Last edited by Nevyn on Sun Jun 10, 2018 8:15 pm; edited 1 time in total
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
One annoying thing about using '<' and '>' is that you have to turn off HTML code in your posts on this forum to get them to show up correctly.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
One problem with these new commands is that they can't be used to directly fan out from a carousel position. You need some other chain to sit between the root atom and the fan. I will need to introduce another fan command that is like fan, but limits it to a half circle. In other words, it would just fan out all items in the Chain Array and not use one of them to focus on.
I can't think of a good name for it at the moment. Maybe fanAll or fanFrom or something like that. Maybe I should change the existing fan to be called radial and then re-use fan to just fan out all items in the array.
I'll think of something, but suggestions are always welcome.
I can't think of a good name for it at the moment. Maybe fanAll or fanFrom or something like that. Maybe I should change the existing fan to be called radial and then re-use fan to just fan out all items in the array.
I'll think of something, but suggestions are always welcome.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
I just had a look at the Methane paper and MBL still can't model that one. I'll have to introduce a new operator, like the flip operator, but it turns the atom onto its side. More to the point, it needs to stop the existing functionality of turning an atom if it only contains a core proton stack and nothing else. This was needed because all atoms a oriented along the north/south axis, but if the atom only has a core, then it is oriented along the X or Z axis, not the Y, so the MBL Visualizer would turn them to be along the N/S axis. For Methane, we need the H atoms to be E/W, not oriented along the N/S axis.

I really don't like this ability of atoms to re-arrange their protons. Very hard to work with in the code. I don't see any rhyme or reason to it. You have to know too much about the molecule to figure out how it might be configured. I want to work from the other direction. I want the underlying principles to guide the configuration. Neodymium is another example that changes in molecules but some protons go up and some go down. What kind of charge field could do that? It just feels like the modeler is deciding that they want more charge channeling somewhere, so they just take protons from somewhere else and put them there. Quite literally the end justifying the means. I know that we have to work that way to figure these things out, but once we do, we should strive to make it work from the other direction. /rant

I really don't like this ability of atoms to re-arrange their protons. Very hard to work with in the code. I don't see any rhyme or reason to it. You have to know too much about the molecule to figure out how it might be configured. I want to work from the other direction. I want the underlying principles to guide the configuration. Neodymium is another example that changes in molecules but some protons go up and some go down. What kind of charge field could do that? It just feels like the modeler is deciding that they want more charge channeling somewhere, so they just take protons from somewhere else and put them there. Quite literally the end justifying the means. I know that we have to work that way to figure these things out, but once we do, we should strive to make it work from the other direction. /rant
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
I was thinking along the same lines the other day while trying to use MBL to model C21H30O2, known widely as THC. It was just for fun, but I realized it would take a lot more work on my end to "describe" it in MBL, since I've only just dabbled in it so far. So the molecule came out basically a long, obscene string instead of the shape it's described to us as traditionally.
But that's not your fault - it's mine. I didn't take the time to learn MBL better to make it work. I'll try again soon.
But that's not your fault - it's mine. I didn't take the time to learn MBL better to make it work. I'll try again soon.
Jared Magneson- Posts : 525
Join date : 2016-10-11
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Jared Magneson wrote:I was thinking along the same lines the other day while trying to use MBL to model C21H30O2, known widely as THC. It was just for fun, but I realized it would take a lot more work on my end to "describe" it in MBL, since I've only just dabbled in it so far. So the molecule came out basically a long, obscene string instead of the shape it's described to us as traditionally.
But that's not your fault - it's mine. I didn't take the time to learn MBL better to make it work. I'll try again soon.
I was thinking of somehow rendering the MBL in a matix style dataset just so machine learning could be applied. How to render each molecule in a table or JSON string while keeping fidelity to particular properties?
How to create a data warehouse/data set layout according to the C.F.? JSON helps a lot but kind hides using structures for other mapping (MBL to traditional Chem Notation-Electron bonding). Ultimately, it would be cool to set the Electron bonding notation and have a ML routine map it back to Mathis with high probability.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
What do you mean by a matrix style dataset? Random green characters flowing down the screen? Probably not, but if you mean to extract the transform matrices for each proton, neutron and possibly electron, then that is feasible. They could be grouped into atoms, sub-grouped into stacks, etc. I don't know what you would need to get it to work with a machine learning system. But I could easily generate some JSON like this:
That really only gives you the positions of things. I imagine that you would have to implement some rules in the learning system for it to make sense but I have no working knowledge of them, so I can't say too much.
- Code:
"molecule": {
"atoms": [
{
"symbol": "He"
"stacks": [
{ "type": "Proton", "position": [X,Y,Z], "rotation": [X,Y,Z] }
{ "type": "Neutron", "position": [X,Y,Z], "rotation": [X,Y,Z] }
{ "type": "Neutron", "position": [X,Y,Z], "rotation": [X,Y,Z] }
{ "type": "Proton", "position": [X,Y,Z], "rotation": [X,Y,Z] }
,
...
]
}
,
...
]
,
....
}
That really only gives you the positions of things. I imagine that you would have to implement some rules in the learning system for it to make sense but I have no working knowledge of them, so I can't say too much.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Nevyn wrote:What do you mean by a matrix style dataset? Random green characters flowing down the screen? Probably not, but if you mean to extract the transform matrices for each proton, neutron and possibly electron, then that is feasible. (YES!)They could be grouped into atoms, sub-grouped into stacks, etc. I don't know what you would need to get it to work with a machine learning system. But I could easily generate some JSON like this:
...snip of code block
That really only gives you the positions of things. I imagine that you would have to implement some rules in the learning system for it to make sense but I have no working knowledge of them, so I can't say too much.
Sorry Nevyn, lately I've been looking at DNA strands and numbered positions for them. I guess, I'm looking for almost an extremely wide table representation of this to use for querying. I'm going to have to look at maybe converting JSON to SQL to try and parse elements into a queryable form. Lazily trying to get things arranged for running R/Python ML algorithms on them. I could possibly scrape an atomic arrangement from Wikipedia for certain molecules and try to rebuild them in the MLB engine. https://upload.wikimedia.org/wikipedia/commons/thumb/3/34/Dichloroacetic-acid-2D-skeletal.png/165px-Dichloroacetic-acid-2D-skeletal.png (an anti-cancer nearly free drug that I'm trying to figure out why this works so well using Miles' C.F. --- researchers really don't know the action of it in vivo and have admitted to seeing the positive effects but not knowing why it is so effective...). These are just rambling thoughts on this approach.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Ahh, DNA. Yes, I've had a go at building the DNA building blocks (GATC) before (many years ago) and they are complicated, for sure. It would be an interesting exercise in MBL. In the very least, it is a hell-of-a-lot easier than how I was doing it back then. You should start a new thread in the Chemistry section for it and we can at least work through the molecular structure of them.
With respect to a database, I have one and you are welcome to it, but I don't know if it will be exactly what you want. I designed a data model to store enough information for me to generate the models for AtomicViewer. I wasn't thinking about querying it for atomic information which I would probably design differently. However, it might be possible to use this database in order to generate a new one with higher level information. It really depends on your needs.
I had not thought about a database populated by MBL expressions before and it seems like an easy thing to do. What I mean is that I create an MBL Renderer that writes or executes the SQL to populate the database. Instead of a renderer that creates 3D models, this one generates SQL for a certain data model. I just need to know either what that data model is, or what it has to accomplish so that I can create it. The SQLRenderer is fairly easy to write unless the data model is too far removed from its concepts.
I might have a go at designing an almost direct representation of MBL as a database schema. It will pretty much be a database version of the Abstract Syntax Tree I've talked about above. Storing the raw data that an MBL expression declares. I'm not sure how useful that will be, but it might be a good start to other things. In the very least, it is easy to do.
With respect to a database, I have one and you are welcome to it, but I don't know if it will be exactly what you want. I designed a data model to store enough information for me to generate the models for AtomicViewer. I wasn't thinking about querying it for atomic information which I would probably design differently. However, it might be possible to use this database in order to generate a new one with higher level information. It really depends on your needs.
I had not thought about a database populated by MBL expressions before and it seems like an easy thing to do. What I mean is that I create an MBL Renderer that writes or executes the SQL to populate the database. Instead of a renderer that creates 3D models, this one generates SQL for a certain data model. I just need to know either what that data model is, or what it has to accomplish so that I can create it. The SQLRenderer is fairly easy to write unless the data model is too far removed from its concepts.
I might have a go at designing an almost direct representation of MBL as a database schema. It will pretty much be a database version of the Abstract Syntax Tree I've talked about above. Storing the raw data that an MBL expression declares. I'm not sure how useful that will be, but it might be a good start to other things. In the very least, it is easy to do.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Cr6, what databases do you like to work in? I use MySQL at home, but often use Oracle, SQLServer and Postgres at work. I have also used SQLite and Apache Derby for small projects. The reason I ask is that writing DDL to populate a schema requires some way of saving row IDs so they can be used in child tables. Each database tends to do it differently. For MySQL I use session variables which are really nice to use. For example, I might do something like this:
The generated script is then run inside your favorite database app such as MySQL Workbench or Oracle SQLDeveloper or PGAdmin, etc.
- Code:
-- get the largest ID for a table and store it in a session variable
select (@chainId:=max(id)) from mbldb.chain;
select (@chainElementId:=max(id)) from mbldb.chain_element;
-- Create a chain
set @chainId := @chainId + 1;
insert into mbldb.chain (id, is_array, type) values (@chainId, 'N', NULL);
-- Create element for the chain.
set @chainElementId := @chainElementId + 1;
insert into mbldb.chain_element (id, chain_id, symbol, is_flipped, rotate, north, south, ordinal) values (@chainElementId, @chainId, 'O', 'N', 0, NULL, NULL, 1);
The generated script is then run inside your favorite database app such as MySQL Workbench or Oracle SQLDeveloper or PGAdmin, etc.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Hey Nevyn, I actually work in SQL Server 2012+ mostly but do work in Oracle/MySQL to some extent. SQL Server 2016+ stores JSON natively so I would be looking at this for making a tabular version of Molecules. This was just a thought on doing ML on tables/tabular forms of the C.F. MBL if at all possible. I just have hunch that properties could be derived from it for automating structures. Might just be too much imagination at this point as well.
Last edited by Cr6 on Sun Jul 15, 2018 1:47 am; edited 1 time in total
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Hi Nevyn, I'm kinda stuck here (diving into MBL a bit) and thought posting here might be helpful for future references and newcomers too. My molecule so far:
ring w 45:(H)6{OH}

My question is, how do I get that Hydrogen on the end of the Oxygen to "turn" and face the O's charge field? I can't seem to get it to rotate on the proper axis. I tried:
ring w 45:(H)6{OH*90}
...and it rotates funny. What am I doing wrong?
Ultimately, I'm trying to generate THC obviously. Hope that doesn't deter anyone from the conversation or helping me out a little, though I understand if some people don't want to touch the topic.
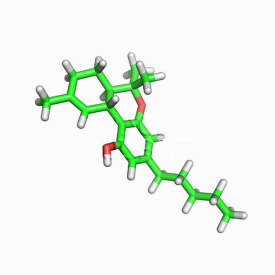
ring w 45:(H)6{OH}

My question is, how do I get that Hydrogen on the end of the Oxygen to "turn" and face the O's charge field? I can't seem to get it to rotate on the proper axis. I tried:
ring w 45:(H)6{OH*90}
...and it rotates funny. What am I doing wrong?
Ultimately, I'm trying to generate THC obviously. Hope that doesn't deter anyone from the conversation or helping me out a little, though I understand if some people don't want to touch the topic.

Jared Magneson- Posts : 525
Join date : 2016-10-11
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
It wouldn't turn, it should be closer and next to the proton in the Oxygen atom. It is a limitation of the MBL Renderer. It doesn't push the atoms together, it just sits them right next to each other.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
You've picked a pretty difficult molecule to work with. There are bonds in that that I am not sure MBL can handle yet. Those connected rings are an issue, for sure. If you have trouble, just call out, because it might be MBL that is the problem.
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Yeah, I mean I knew it was a beast and perhaps I overreached here to start with. I'm gonna back up and work on a few much less complex molecules and learn the language better. It's really cool and I'm thankful you spent the time to take it even this far!
Jared Magneson- Posts : 525
Join date : 2016-10-11
 Re: Molecular Bonding Language
Re: Molecular Bonding Language
Thanks, Nevyn. It's obviously not critical, I was just trying to figure out from a mechanical perspective why this molecule is so unique. I guess it just has a lot of loose ends to bond with other substances? I mean, there's nothing "magical" about Oxygen, Hydrogen, and Carbon on its own. But it's been called among the most complex natural molecule occurring in plants, so I always wondered if that was true.
Just an aside, nothing to worry about here. It's fun to study and MBL really helps visualize things.
Just an aside, nothing to worry about here. It's fun to study and MBL really helps visualize things.
Jared Magneson- Posts : 525
Join date : 2016-10-11
Page 2 of 2 •  1, 2
1, 2
 Similar topics
Similar topics» Miles Periodic Table with Standard Periodic Table reference
» Atomic Modeling Language
» Spinning Particle Language
» Natural Language Processing (NLP) and newer algorithms
» Hypervalent bonding controversy out for the electron count?
» Atomic Modeling Language
» Spinning Particle Language
» Natural Language Processing (NLP) and newer algorithms
» Hypervalent bonding controversy out for the electron count?
Page 2 of 2
Permissions in this forum:
You cannot reply to topics in this forum