Miles Periodic Table with Standard Periodic Table reference
Page 3 of 13 •  1, 2, 3, 4 ... 11, 12, 13
1, 2, 3, 4 ... 11, 12, 13 
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
LongtimeAirman wrote:.
https://github.com/LtAirman/ChargeFieldTopics
Well Cr6, it could sure use plenty of improvements, nevertheless, I declare Atom Builder Version 1 well started.
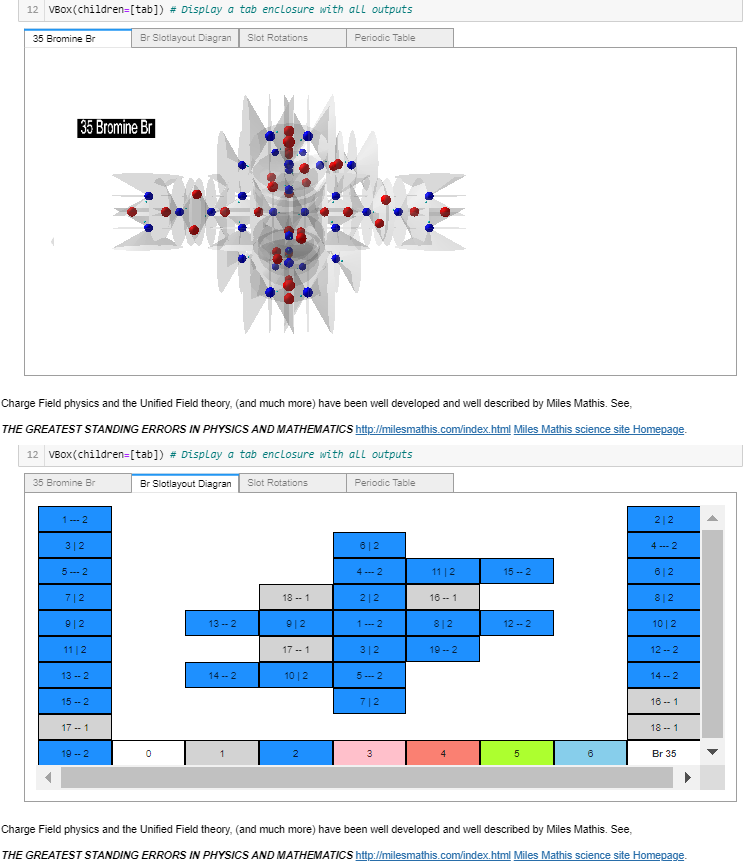
I racked my brains and came up with what I believe is a suitable finishing touch. Just before the notebook’s final reference cell, another widget containing: the rendered atom, its slotlayout diagram, the rotation controls, and a periodic table (with non-functioning buttons – to prevent changing the selected values). All in a single location selectable by tab, as shown in these four images.
I'm spending some time appreciating it.
.
Ace work LTAM! Seriously! I'm just trying to think of algorithimic short cuts right now for rendering. It is looking better and better. The tabs is an excellent approach. It kind of gets me thinking though that if a proper AI/ML type algo was created...it could cut across Standard Bonding to a Slot Layout version? Basically, given these inputs and bonding for atoms this molecule (with standard theory) could render a Slot Layout version (Miles' style). What is built now is good enough. Great code!
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
elements = ((' 1 Hydrogen H ',1),(' 2 Helium He ',2),(' 3 Lithium Li ',3),(' 4 Beryllium Be ',4),
(' 5 Boron B ',5),(' 6 Carbon C ',6),(' 7 Nitrogen N ',7),(' 8 Oxygen O ',
(' 9 Fluorine F ',9),(' 10 Neon Ne ',10),(' 11 Sodium Na ',11),(' 12 Magnesium Mg ',12),
(' 13 Aluminum Al ',13),(' 14 Silicon Si ',14),(' 15 Phosphorus P ',15),(' 16 Sulfur S ',16),
(' 17 Chlorine Cl ',17),(' 18 Argon Ar ',18),(' 19 Potassium P ',19),(' 20 Calcium Ca ',20),
(' 21 Scandium Sc ',21),(' 22 Titanium Ti ',22),(' 23 Vanadium V ',23),(' 24 Chromium Cr ',24),
(' 25 Manganese Mn ',25),(' 26 Iron Fe ',26),(' 27 Cobalt Co ',27),(' 28 Nickel Ni ',28),
(' 29 Copper Cu ',29),(' 30 Zinc Zn ',30),(' 31 Gallium Ga ',31),(' 32 Germanium Ge ',32),
(' 33 Arsenic As ',33),(' 34 Selenium Se ',34),(' 35 Bromine Br ',35),(' 36 Krypton Kr ',36),
(' 37 Rubidium Rb ',37),(' 38 Strontium Sr ',38),(' 39 Yttrium Y ',39),(' 40 Zirconium Zr ',40),
(' 41 Niobium Nb ',41),(' 42 Molybdenum Mo ',42),(' 43 Technetium Tc ',43),(' 44 Ruthenium Ru ',44),
(' 45 Rhodium Rh ',45),(' 46 Palladium Pd ',46),(' 47 Silver Ag ',47),(' 48 Cadmium Cd ',48),
(' 49 Indium In ',49),(' 50 Tin Sn ',50),(' 51 Antimony Sb ',51),(' 52 Tellurium Te ',52),
(' 53 Iodine I ',53),(' 54 Xenon Xe ',54),(' 55 Caesium Cs ',55),(' 56 Barium Ba ',56),
(' 57 Lanthanum La ',57),(' 58 Cerium Ce ',58),(' 59 Praseodymium Pr ',59),(' 60 Neodymium Nd ',60),
(' 61 Promethium Pm ',61),(' 62 Samarium Sm ',62),(' 63 Europium Eu ',63),(' 64 Gadolinium Gd ',64),
(' 65 Terbium Tb ',65),(' 66 Dysprosium Dy ',66),(' 67 Holmium Ho ',67),(' 68 Erbium Er ',68),
(' 69 Thulium Tm ',69),(' 70 Ytterbium Yb ',70),(' 71 Lutetium Lu ',71),(' 72 Hafnium Hf ',72),
(' 73 Tantalum Ta ',73),(' 74 Tungsten W ',74),(' 75 Rhenium Re ',75),(' 76 Osmium Os ',76),
(' 77 Iridium Ir ',77),(' 78 Platinum Pt ',78),(' 79 Gold Au ',79),(' 80 Mercury Hg ',80),
(' 81 Thallium Tl ',81),(' 82 Lead Pb ',82),(' 83 Bismuth Bi ',83),(' 84 Polonium Po ',84),
(' 85 Astatine At ',85),(' 86 Radon Rn ',86),(' 87 Francium Fr ',87),(' 88 Radium Ra ',88),
(' 89 Actinium Ac ',89),(' 90 Thorium Th ',90));
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
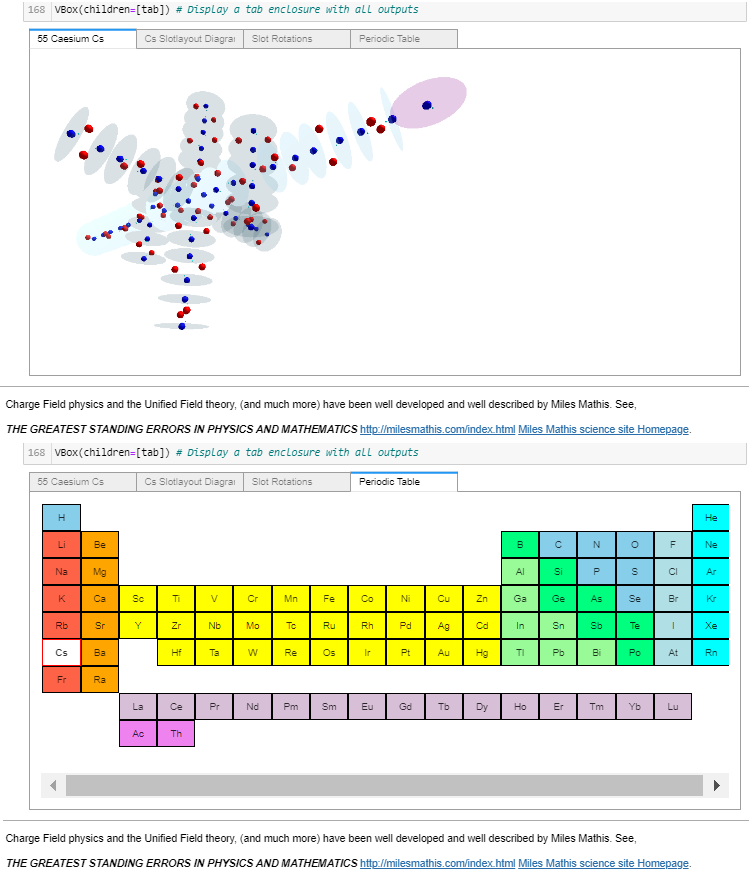
Atom Builder (AB) now displays the selected atom in the tab’s periodic table. After making that change a couple of days ago, I’ve stopped working on AB.
Our next task is to display molecules. Basically coming up with the bonding rules that allow us to join - or not, a specified set of atoms together. The most important rule may be that the proton stacks at the bonding sites between adjacent atoms must join together in either a single parallel stack (of up to six protons) or join as two adjacent orthogonal stacks.
I believe atoms will usually string together along their main vertical columns – displayed horizontally for convenient viewing. I don’t see how adding an element’s 19 slots ‘horizontally’ or ‘vertically’ would make any difference in rendering speed. I thought we’d agreed that the best way to speed things up would be by replacing AB’s 19 slot i-loops with a data set’s 19 slot lookup values.
Toward that end, I’ve added the two Cr6-Elements .json and .csv files from Nevyn’s Atom Viewer project to the AB file folder to be accessed by a second notebook file I’ve started calling Molecule Builder (MB). I’ve been studying you excel document MathisAllElementsv30B.xlsx more closely as well as reviewing u-tube tutorials on the subject of reading, writing and parsing data files, substituting your file in place of the tutorial examples when possible.
Everything appears straight forward enough.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
I’ve begun mBuilder, at least in my own mind, although I’m still working through a tutorial or two.
Several items, mostly csv file changes:
Here’s the first cell’s markdown code intro.
The original Cr6-Elements.csv (comma separated value) data file contains 3,016 rows and 17 columns, allowing for up to more than 150 atomic elements. The numerical order of the elements larger than 118 is gone, and only the first 90 elements contain Slotlayout diagrams, so as with AB, I started by eliminating all the elements larger than 90.
## Molecules
mBuilder will render a 3D charge field model of molecular matter from atoms selected by the user.
That's the plan anyway. At first I'll try making atoms. Molecules is Version 2 of Atom Builder (AB), replacing AB's 19 i-loop slots with a 19 slot csv data file. The expected faster renderings and efficiencies gained should allow us to build multiple atoms in molecular configurations.
We might end up using the json data file instead. I'll try reusing most of AB's code, such as the periodic table. This time around we'll use Cr6's original Slotlayout (SL) diagram.
Molecules is intended to introduce Charge Field theory – see the notebook's end cell's source references.
Four Column headers over each individual element’s slotlayout diagram were blank, so I added: ‘SL1’, ‘SL2’, ‘SL3’, ‘SL4’. Here’s the file’s current header row:
Atomic Number, AtomicSymbol, Element, OrbitalGroup, Protons, Neutrons, GroupNumber, Period, SlotNumber, SlotOrder, CanBind, AlphaType, Slot Layout, SL1, SL2, SL3, SL4
Each ‘Element’ value, (or every atomic name), was followed by a space and then a comma. That produced a four character suffix - A(with a top carrot^)xa0. I removed the space after every element name.
Each element should have just 19 rows, but 6 elements (Berylllium, Boron, Carbon, Nitrogen, Oxygen, and Fluorine) had more than 19, I suppose due to the unusual small atom slot 1 proton counts. I removed a total of 13 excess rows. Cr6-Elements now contains a header row + 19*90 remaining record rows. 1,711 rows by 17 columns.
My current task is to recreate the elements tuple array from AB using the Cr6-Elements.csv file.
elements = ((' 1 Hydrogen H ',1),(' 2 Helium He ',2), … ,(' 90 Thorium Th ',90)).
That particular effort's current code and output:
- Code:
import csv
#filename = '../data/tBoron.csv'
filename = '../data/Cr6-Elements.csv'
with open(filename) as csv_file:
csv_reader = csv.reader(csv_file, delimiter=',')
line_count = 0
for row in csv_reader:
if line_count == 0:
print(f'Column names are {", ".join(row)}')
line_count += 1
else :
if line_count % 19 == 0 :
print(f'\t{row[0]} {row[2]} {row[1]}')
line_count += 1
print(f'Processed {line_count} lines.')
#Output:
Column names are Atomic Number, AtomicSymbol, Element, OrbitalGroup, Protons, Neutrons, GroupNumber, Period, SlotNumber, SlotOrder, CanBind, AlphaType, Slot Layout, SL1, SL2, SL3, SL4
1 hydrogen H
2 helium He
3 lithium Li
...
88 radium Ra
89 actinium Ac
90 thorium Th
Processed 1711 lines.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Attempting to recreate AB’s elements=((,),…,(,)) tuple series (I’ve wrongly referred to as tuple array before) this time with pandas, a powerful and popular python library I recently found out about.
Little to no tangible project progress I’m afraid Cr6.
After making great efforts at rather small file changes and working tutorials, studying the python csv library, I found pandas could read the .csv files without errors. The csv code I print() posted last time just mimicked elements’ tuples, the above image shows how pandas may be used to create a true atomic tuple (‘Element string label’, and AtomicNumber value). I’d still need to add the atomic symbol somehow. Then the elements tuple array (x19 copies of each element – 18 too many) would be an internal dataframe csv read file column series. So then what do I do? At this point, I don’t believe adding the tuple series to Cr6-Elements.csv would slow anything down, so I’ll probably just add it each 19th row, the same row containing each atom’s Slot_Layout title/starting row.
Cr6-Elements.csv needs some changing.
1. Most important, while each SL diagram indicates the number and orientation of the protons in each atom’s occupied slot configuration, the actual proton positions are missing and needs to be added; ideally as functions of the atomic particles’ (protons/neutrons/electrons’) proton radius separation distances as used in atom builder. That data is currently available in AB’s 19 slot loops and proStack function.
2. Lower priority, while I believe the SL can, it currently needs to diagram extra non-alpha, neutron counts as well as their orientations and calculated positions. Such would be needed for accuracy and isotopes.
Like so many things in this project, data files and data structures are new to me, I need to keep learning. I’m far from adequately understanding csv, and haven’t even begun reviewing json yet, but pandas and also possibly numpy(another python database and data science library) seems to be easier and more promising, so I’ve started reviewing/studying pandas tutorials. Luckily pandas.pydata.org has plenty of examples, they’re even built into jupyter notebook’s library documentation – accessible by cursor position when typing pandas code, using the shift tab keys.
https://pandas.pydata.org/docs/user_guide/10min.html#min
Please feel free to redirect me as you see fit. It takes a while sometimes but I'll keep trying.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
If you want to take a break and try something completely different, take a look at these clustering algos in the sklearn library. https://machinelearningmastery.com/clustering-algorithms-with-python/ It might be possible to build up models for a Miles' periodic table similar to what the Chinese did with the old periodic table. This library should work with jupyter: https://github.com/microsoft/FLAML
Looking into Panda-Python's "map" function might useful as well.
https://towardsdatascience.com/apply-function-to-pandas-dataframe-rows-76df74165ee4
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
take a break and try something completely different
Cr6, thanks for the break. In the old days one needed to wait while a program loads, now you may need to wait while the program learns. Those clusters are also described at https://scikit-learn.org/stable/modules/clustering.html They appear to be based on the distances between all the data points, and how well those points are described by those algorithms. I’ve downloaded a lot from github lately, https://github.com/microsoft/FLAML is now bookmarked for future reference. You’ve corrected my attitude about machine learning in the past. Yesterday I received an early xmas present, only one such item a year, which I did not select, Machine Learning with Python for Everyone, by Mark E Fenner. It has a small section on clusters. Something completely different or cosmic coincidence?
“similar to what the Chinese did with the old periodic table”.
I recall you mentioning the “Chinese periodic table” once before, but I don’t remember any details and my local file searches turn up empty. After a short search “in the cloud”, I found something … . Do you mean “Wuxing”?
https://pages.ucsd.edu/~dkjordan/chin/WuuShyng.html
The Five Elements:
An Ancient Chinese "Thinking Machine”.
and

https://en.wikipedia.org/wiki/Wuxing_(Chinese_philosophy)
I can certainly see Wuxing as a charge field philosophy of change. Starting at: up/down, left/right, or front/back directions and the prevailing flows of charge and anticharge (2:1) throughout nature here on earth. The paths to change will primarily be in two of the available five directions, in accordance with the charge or anti-charge flows and the five phase way.Wuxing (Chinese philosophy)
From Wikipedia, the free encyclopedia
Diagram of the interactions between the wuxing. The "generative" cycle is illustrated by grey arrows running clockwise on the outside of the circle, while the "destructive" or "conquering" cycle is represented by red arrows inside the circle.
Wuxing (Chinese: 五行; pinyin: wǔxíng), usually translated as Five Phases, is a fivefold conceptual scheme that many traditional Chinese fields used to explain a wide array of phenomena, from cosmic cycles to the interaction between internal organs, and from the succession of political regimes to the properties of medicinal drugs. The "Five Phases" are Fire (火; huǒ), Water (水; shuǐ), Wood (木; mù), Metal or Gold (金; jīn), and Earth or Soil (土; tǔ). This order of presentation is known as the "Days of the Week" sequence. In the order of "mutual generation" (相生; xiāngshēng), they are Wood, Fire, Earth, Metal, and Water. In the order of "mutual overcoming" (相克; xiāngkè), they are Wood, Earth, Water, Fire, and Metal.[1][2][3]
The system of five phases was used for describing interactions and relationships between phenomena. After it came to maturity in the second or first century BCE during the Han dynasty, this device was employed in many fields of early Chinese thought, including seemingly disparate fields such as Yi jing divination, alchemy, feng shui, astrology, traditional Chinese medicine, music, military strategy, and martial arts. Although often translated as the Five Elements in comparison to Classical elements of the ancient Mediterranean world, the Wuxing were conceived primarily as cosmic agents of change rather than a means to describe natural substances.
If not, oops; never mind, what did you mean by Chinese periodic table?
Panda-Python's "map" function might (be) useful as well
Yes, I’ve see Pandas map function used in a couple of the video examples in the last few days, but haven’t gotten that far working examples in the user guide yet. Its a definite plus in favor of pandas.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
[size=57]Machine learning material properties from the periodic table using convolutional neural networks†[/size]
[size=59]Abstract[/size]
[size=58]In recent years, convolutional neural networks (CNNs) have achieved great success in image recognition and shown powerful feature extraction ability. Here we show that CNNs can learn the inner structure and chemical information in the periodic table. Using the periodic table as representation, and full-Heusler compounds in the Open Quantum Materials Database (OQMD) as training and test samples, a multi-task CNN was trained to output the lattice parameter and enthalpy of formation simultaneously. The mean prediction errors were within DFT precision, and the results were much better than those obtained using only Mendeleev numbers or a random-element-positioning table, indicating that the two-dimensional inner structure of the periodic table was learned by the CNN as useful chemical information. Transfer learning was then utilized by fine-tuning the weights previously initialized on the OQMD training set. Using compounds with formula X2YZ in the Inorganic Crystal Structure Database (ICSD) as a second training set, the stability of full-Heusler compounds was predicted by using the fine-tuned CNN, and tungsten containing compounds were identified as rarely reported but potentially stable compounds.[/size]https://pubs.rsc.org/en/content/articlelanding/2018/sc/c8sc02648c#!divAbstract
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Current Boron data and atomic model image from MathisAllElementsv30.xlsx (and initial Cr6-Elements.csv files). The column headers’ index and labels are: A AtomicNumber, B AtomicSymbol, C Element, D OrbitalGroup, E Protons, F Neutrons, G GroupNumber, H Period, I SlotNumber, J SlotOrder, K CanBind, L AlphaType, M Slot Layout, N SL1, O SL2, P SL3, Q SL4. Boron is one of 90 such 19 row sections, each containing its own 5 row x 11 column slotlayout sub-section containing cells with both proton counts and proton emission orientation. The Boron's dark blue and black colors (representing two alpha protons and single non-alpha protons) are hard to distinguish so they were changed to lighter blue and gray in the SL diagrams. The proton stack color legend is added for clarity. Note Boron’s slot three contains two – not one, non-alpha protons which necessitates some additional data handling detail. I added a boron image from AB as well. I implemented boron’s two non-alpha protons as a pair of alpha protons for convenience. AB also renders single protons the same color as the legend’s three proton stack color instead of gray, since the pink is easier to distinguish from the rendered blue (too close to gray) alphas. Your tolerance is appreciated.
Status update. Am working through hundreds of code examples, about halfway through the ‘Essential basic functionality’ section of the pandas user guide at https://pandas.pydata.org/docs/user_guide/basics.html. Plenty of code I need to see used before attempting to do so myself. I can only concentrate on it about two or three hours a day.
As previously mentioned, each atom’s proton stack positional information needs to be added to Cr6-Elements.csv, along with other small changes. Once those changes are in place, I imagine the data file will be used almost exclusively for table look-ups, a simpler use than the great majority of pandas user guide code examples.
I read “Machine learning material properties from the periodic table using convolutional neural networks”, and had some difficulty understanding how it worked. I can certainly appreciate that new machine learning programs are able to make extremely accurate material property predictions, based on the initial input data and the shape of the standard periodic table alone. There's no question specially designed machines can “perceive” patterns in nature better than we can. Note that the ball and stick 3x3x3 27 atoms in a cubic configuration “Crystal structure of an L21 full-Heusler compound” looks like a somewhat over-simplified pre-condition. As I see it, the problem lies in describing the data results properly. As Miles has repeatedly pointed out, throughout scientific history, the great thinkers, such as Newton or Einstein, usually created formulae more in accordance with observation than their theories did in explaining the data – more heuristic than theoretical. Those great thinkers missed the fact that they were taking their data from a dual, opposing, unified gravity/charge field. Someone should give the machines that fact and a charge field atomic model to see what they predict next.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

The column headers’ index and labels are: A AtomicNumber, B AtomicSymbol, C Element, D OrbitalGroup, E Protons, F Neutrons, G GroupNumber, H Period, I SlotNumber, J SlotOrder, K CanBind, L AlphaType, M Slot Layout, N SL1, O SL2, P SL3, Q SL4.
Status. Still reviewing pandas’ user guide.
More thoughts on changing the data file.
1. Column J, SlotOrder, has a single value of 1 repeated 19x90 rows over all Cr6-Elements.csv data. It may be eliminated. Or reused to hold a slot’s proton or neutron, position or orientation or alpha status data.
2. For any given element, 13 of the total 17 columns have the same data repeated for all the element’s 18 rows. For Boron: A AtomicNumber = 5; B AtomicSymbol = B; C Element = Boron; D OrbitalGroup = [He]; E Protons = 5; F Neutrons = 6; G GroupNumber = 13; H Period = 2; and J SlotOrder = 1.
3. The SL table section. I see the SL as that atom’s 3D atomic projection onto the excel grid, showing the atom’s unique proton stacks’: count, orientation and alpha-status. Note that the color-coding is for human benefit, and will not (as far as I know) be part of any .csv file based dataframe operations. In fact, for the datafile data’s sake, each slots’ proton: count, orientation and alpha status should have its own data columns. The dataframe shouldn’t need to read data from the SL diagram, the SL diagram should be built from proton columnar data.
The SL’s 5 cols (not rows) by 11 rows (not cols), with four blank rows represents the greater emission direction distances of top and bottom slots 14 and 15, toward their adjacent vertical column slots 4 and 5. That distancing however is not applied between slots 2 and 3 and their vertical neighbors, or between the hook positions and the main column slots they are adjacent to; no doubt each for their own SL visual balancing reasons. In AB those 4 blank rows were skipped, the SL is 5 cols by 7 rows and remains an equally valid atomic representation.
4. There’s good reason to eliminate the four .csv SL blank rows; to allow room for a second SL diagram - for neutrons. Two times 11 is 22 rows, larger than each element’s current 19 rows. There’s no such problem in adding a neutron SL when each SL is only 7 or 9 rows. The neutron SL diagram, like the proton, should be built from neutron columnar data not currently present, which would also need to be added.
5. Drop every element’s first row, col “M“, Slot Layout” title string object. One should not mix data types in a given column in a pandas data frames. As is, column M contains several data types: 1. Slot numbers – 10 and 11; 2. Slot’s 10 and 11 proton emission “directions” - horizontal or vertical; 3. Slot’s 10 and 11 proton counts (using color); and 4. The element’s SL title string. Column M’s header also contains the SL title string which makes repeating the same title for each element redundant.
6. As previously mentioned, in addition to the slot number, we need the calculated position for an occupied slot’s proton – or one or two neutrons. Convert column J, SlotOrder to SlotCoord, the slot’s central coordinates, good for any or all objects. That data would be an (x,y,z) 3d Vector. I’ve not yet run across coordinates in my pandas review, or the single SlotCoord column can be three columns, such as SlotCoordx, SlotCoordy and SlotCoordz.
7. With 1. the proton and neuton data separation and column additions mentioned above; 2. previous discussion of the Elements tuple series used for the periodic table buttons; and 3. the location of the slot’s object. A final “cleaned up” Cr6-Elements.csv file can end up with 22 columns: A AtomicNumber, B AtomicSymbol, C Element, D AtomicTuple, E OrbitalGroup, F Protons, G POrient, H PCount, I PAlpha, J Neutrons, K NOrient, L NCount, M NAlpha, N GroupNumber, O Period, P SlotNumber, Q CanBind, R SlotCoord, S SL1, T SL2, U SL3, V SL4.
Is all that agreeable to you???
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Last edited by Chromium6 on Mon Dec 20, 2021 8:37 pm; edited 1 time in total
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
https://spacy.io/ There are a lot of python packages out there. So Spacy is a transformer-based pipeline program application that can work with convolutional neural network programs (CNNs)? Deep learning sounds terrifically complicated. I’m at my limit dealing with your Cr6-Elements.csv file; I've made the changes listed below, let me know if you want any other changes.
Am currently in the middle of the pandas user guide’s “Indexing and selecting data” section. I’m learning slowly, and am anxious to get back to coding.
Changes to Cr6-Elements and tBoron:
1. Trimmed excess spaces:
a. In every row after every Element name.
b. In the 7 x 90 rows preceeding (left side) each SL diagram orientation hypen (-) chartacter.
c. In SL diag From "| 3" to "| 3", (52 rows).
d. Column M header, removed space from Slot Layout changing to SlotLayout.
2. SL diagram cleanup, standardizing to orientation character (| or -) single space “SlotNumber”.
3. Removed 90 occurrences, (at each element’s SL diagram), of “Slot Layout”.
4. Replaced column SlotOrder with three slot position (a position, not a vector) columns: SlotX, SlotY and SlotZ. All values are currently zero.
5. Added SlotP – the number of protons in that slot. All values are currently zero.
6. Added SlotN – the number of neutrons in that slot. All values are currently zero.
7. Added SlotOrien – the emission orientation of that slot. All values are H or V for horizontal and vertical as per the SL diag.
8. Added AtomTup, i.e. "(' 90 thorium Th ', 90)".
The column headers are now: A AtomicNumber, B AtomicSymbol, C Element, D AtomTup, E OrbitalGroup,F Protons, G Neutrons, H GroupNumber, I Period, J SlotNumber, K SlotP, L SlotN, M SlotOrien, N AlphaType, O SlotX, P SlotY, Q SlotZ, R CanBind, S SlotLayout, T SL1, U SL2, V SL3, W SL4
Still not sure how I’ll handle AlphaType; as a boolean or as a number less than or equal to SlotP?
There’s still the occasional odd characters in the SL diag portion of the read .csv files that haven’t caused any errors. I use LibreOffice Calc in place of excel, and make most file edits with Notepad++. As in AB, the program will output its own SL diagram, relying on column data and not each element’s csv SL diagram. Since SlotOrien, SlotX, Slot4Y, SlotZ and AlphaType applies to both protons or neutrons, I realize a second SL diagram for neutrons isn’t necessary. Each element’s csv SL diagram dimensions remain the same.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Cr6 wrote. ‘Just to clarify the spacy python library is often used to translate languages, syntaxes grammer from one human language to another. Looking for something similar with Miles' table and the classic periodic table. It may require adding more direction-position-spin-angle details to Miles' atoms. Just a thought.’
Airman. Thanks for sharing your thoughts, I usually need plenty of time to consider them before arriving at a decent understanding. Along that line I had a bit of an aha moment when I read the following quote *.
Hierarchical / Multi-level indexing is very exciting as it opens the door to some quite sophisticated data analysis and manipulation, especially for working with higher dimensional data. In essence, it enables you to store and manipulate data with an arbitrary number of dimensions in lower dimensional data structures like Series (1d) and DataFrame (2d).
At first I thought of each element x in terms of up to 19 slot rows by 23 cols, and we could do without multi-indexing. Now I tend to see slots and their associated protons strictly as dimensional values of orthogonal directions and magnitude. This is just pandas, not deep learning, but along with tools like multi-indexing, pandas appears to have the ability to do just the sort of data analysis you’re looking for.
* From the pandas user guide’s “MultiIndex / advanced indexing” section. Hierarchical indexing (MultiIndex).
https://pandas.pydata.org/docs/user_guide/advanced.html
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Wasn't aware that pandas had Hierarchical / Multi-level indexing built in. Looks promising for adding relationship properties for C.F. style bonding. Reminds me of graph databases a bit as well.
Miles has done a good job of indicating why certain atoms can bond while others can't with his models and calcs. While traditional physics-chemistry can kind of define relationships for bonding...Miles with a good table could allow ML to help shortcut discovery for what is allowed and the properties for why it is allowed...at least for prime cases. Basically as simple molecules like CO2 or CO or NaCl get defined as allowed via slots-C.F. direction and weights(?)....the Miles' version can say if that bonds on his numbers or due to a special case like the disk angle or something. Tbh....most of the software for atomic modeling looks pretty crude with traditional bonding theory.
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
I read you Cr6. I haven't collected Miles' bonding rules, have you? We doubtless need to quantify a few things which will get difficult. On the other hand, things like the alpha model (He) and charge flow across orthogonal proton stacks seem central, sufficient and simple enough.
Halfway through “Merge, join, concatenate and compare”, the 7th in a list of 26 sections. Good thing they provide so many examples, otherwise I wouldn’t have had any real understanding of the subject. The tutorials I first reviewed were skimmed from portions of the pandas user guide.
For single atoms I’m sure I wouldn’t need to read the entire guide, but we want to display more than a single atom. My current thinking, feel free to correct me, each atom is an independent dataframe. Bonding will involve multi-indexing and may in fact be perfectly analogous/applicable to a pandas built-in joining or concatenating operations – left, right, (up/down), intersection, union, cross. Each element’s dataframe will need to include the set of up to 6 (: front/back left/right, top/bottom) extremity/bonding points, along with their spin information to ensure compatible bonding with another atom’s bonding point. Plus the changes to both atoms’ proton configurations as a result of bonding - like shift to one side or the other. Bonding must be a function of atomic charge flows and c.f. rules that may or may not yet be codified - Slap me if I sound delirious. I suppose all that and more must be contained in each dataframe.
I’ll keep working through the examples and proceed after I have a better understanding of the alternatives.
Again, please pardon the delay, and best wishes, I hope you’re doing well.
https://pandas.pydata.org/docs/user_guide/merging.html
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Set thinking may allow flexibility for infinite Null structures until something actually exists there in a slot for a bond-reflection-temperature-rotation-color-etc...then the rules start to apply. Maybe pandas can help out with showing it. Sorry for the over philsophicalization.
Last edited by Chromium6 on Sat Jan 01, 2022 3:15 am; edited 2 times in total
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Status. Am currently in the “Working with text data” section (the ninth), of the Pandas User Guide.
Set thinking sounds right, it may describe atomic bonding well. I’m certain we won’t need infinite null structures, just a few should do. And while I don’t often think in sets, I guess it’s important to work at good indices and let the computer – in this case pandas, do the rest. Overall, I agree, the charge field atomic model and set thinking is a framework that can no doubt better match existing data and perhaps spark a long overdue revolution in atomic physics.
Best wishes as well LTAM and Lloyd-Team!
Set thinking may allow flexibility for infinite Null structures until something actually exists there in a slot for a bond-reflection-temperature-rotation-color-etc...then the rules start to apply. Maybe pandas can help out with showing it. Sorry for the over philisophilication (-zation?).
Philosophizing
Happy New Year!
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Status update. Continued slow progress, over halfway through the pandas user guide, in the 17th section, (note, the sections are not numbered and subject to change over time) – Computational tools. It’s become much easier to work through the examples and I’m experiencing way less “What the huh?” moments. I’ve yet to see pandas used in a working project and am looking forward to the guide’s final section – Cookbook, hopeful for some additional understanding and insight.
There’s one section I was unable to review, section 15 - Chart visualization *, in which matplotlib.pyplot is imported, allowing pandas’ Series and DataFrames to output graphs, or plots.
- Code:
import matplotlib.pyplot as plt
# The plot method on Series and DataFrame is just a simple wrapper around plt.plot():
Running my copy of the example code, (after re-typing or copy-pasting on my machine, adding commentary, etc), produced no output graphs - and no errors, i.e. Mathplotlib is installed. In the same notebook I then copied code from the corresponding (sans pandas) matplotlib tutorial ** which did produce the correct output graphs. Just the pandas plot() version didn’t work. Since I don’t anticipate needing to create any graphs I skipped that pandas section and will eventually return to it or the mathplotlib page instead.
I’m also halfway through “NumPy: the absolute basics for beginners”
https://numpy.org/devdocs/user/absolute_beginners.html
Am re-reading “How to Build the Elements”. Here’s a paragraph I keep coming back to.
Makes me believe that well aligned, (pole-to-pole) protons and neutrons can be in very close proximity and occasionally bump or collide. How close can they get? Electrons present between the nuclear particles no doubt help prevent some collisions, but stopping them? I would assume the equilibrium between gravity and minimal polar charge emissions (like through charge and and near through charge would determine the minimum separation distance – somewhere outside the particle/collision radius; but I’m not sure.Miles wrote. I have shown two things that explain this without a strong force. One, charge does not exist in the nucleus in the way we have been told, so it doesn't resist gravity. This makes gravity much stronger than normal. In other words, we don't have the unified field in all parts of the nucleus, we only have gravity. You will understand what I mean by this once you study some of the nuclear diagrams. Two, I have shown that gravity is much much stronger at the nuclear level than is currently thought. Particle physicists have told us there is no gravity there, but that is a lie or a mistake. Gravity is not only present, it is enhanced.
I’ll also be re-reading several other Miles' papers, Diatomic Hydrogen comes to mind, and an alpha model paper - don't recall a title. And then some.
Not unreasonable at all. Somehow, an “endless universe” sounds more reasonable to me than an “infinite universe”. Not sure of your original meaning I should have added a smiley face to “just a few should do” - infinite null structures - that is. I admit I’m prejudiced against casually casting infinite structures about, null or not. On the other hand, it’s certainly sounds like reasonable 3D thinking to me.Cr6 wrote. Yeah... it was pretty late when I posted the above. I was for some reason thinking of "empty space" as Null structures. Like if the universe in size is truly endless...it may not be a unreasonable thought.
You’re a veritable font of python libraries, Cr6. After a brief review I gather graphframes is a logical extension or expansion on the notion of dataFrames. Say a dataFrame contains all the nodes or members of a set, then the graphFrame is a “related” frame which contains all the relationships between the dataFrame nodes. GraphFrames appear to be intended to speed up “big data” (10^9 – 10^12), billions or more objects, networking type computations and queries. GraphFrames set thinking – machine learning - may be suitable for a possible version 3 – many atoms.Cr6 wrote. In addition to Hierarchies in Pandas this another library to throw in the bucket: https://graphframes.github.io/graphframes/docs/_site/quick-start.html .
*
https://pandas.pydata.org/docs/user_guide/visualization.html
**
https://matplotlib.org/stable/tutorials/introductory/pyplot.html#sphx-glr-tutorials-introductory-pyplot-py
http://milesmathis.com/index.html
315. How to Build the Elements. http://milesmathis.com/nuclear.pdf Explaining the periodic table, with nuclear diagrams. 71pp.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
I think you are looking at this accurately. At the end of the day what is the best relational way to represent "how can a carbon atom 'bond' with an oxygen atom based solely on properties of both atoms according to Miles' structures? 2-D style tables can sort of do it but graph databases-frame may allow more properties for linking instead of just wide dataframe columns. It is a question of why or why not can these two atoms link or chain with Miles' structures? Graph databases can allow that a bit better in a query. I see the graph vertexes-edges a lot like the charge field alignments. A lot of graph databases can represent social networks based on properties and edge relationships between members.
https://github.com/sjmickiemouse/cs229_molecules.git
http://cs229.stanford.edu/proj2019aut/data/assignment_308832_raw/26627851.pdf
http://infochim.u-strasbg.fr/FC/docs/Intro/FC_DB.pdf
OGB-LSC @ KDD Cup 2021 - Open Graph Benchmark
https://ogb.stanford.edu/kddcup2021/pcqm4m/
here they try to do it with rdkit posted on page one of this thread but it is really basic: https://github.com/snap-stanford/ogb/blob/master/ogb/utils/mol.py#L6
https://jcheminf.biomedcentral.com/articles/10.1186/s13321-019-0362-7
This old paper kind of outlines the failures the had using classical bonding algos. Looking for something much simpler and direct with Miles' table https://almob.biomedcentral.com/articles/10.1186/s13015-014-0023-3
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
The reaction matrix Rm encodes the imaginary transition state (ITS):
https://almob.biomedcentral.com/articles/10.1186/s13015-014-0023-3/figures/3
Hydrogen symmetry problem. Symmetries resulting from interchangeable hydrogens. The figure presents three successive atom assignments within an ITS mapping. Bonds present in I are given in black, bonds to be formed to derive O are dotted and gray. The ITS describes the loss of a hydrogen for the carbon (bond order decrease) and the bond formation between the decoupled hydrogen with the oxygen next in the ITS. It becomes clear that all 4 hydrogens are not distinguishable, which results in 4 possible symmetric ITS mappings.
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Currently in the pandas user guide, section 18 Group by: split-apply-combine .
https://pandas.pydata.org/docs/user_guide/groupby.html
Thanks Cr6, Your links have given me plenty to think about. Starting with the news that at about one degree above absolute zero "flickers" of superconductivity have reported been demonstrated using ABC trilayer graphene. I almost posted a response on the Re: Mathis on Graphene? Any hints? thread.
Airman. I hope. My last chemistry class was forty years ago. I can recognize the 2D and 3D structures, all the computational stuff is new. I'm a bit overwhelmed by ITS mapping, adjacency matrices, distance matrices, bond matrices, connection tables, and much more. Definitely improving my awareness and perspective, helping me organize my thoughts. At first I thought I’d complete my pandas guide review before trying to decide on how to proceed, but your links make it clear that in order to find the “best relational way” to implement bonding I need to review such contemporary computer based molecular chemistry alternatives most of which apparently utilize some graph theory.Cr6 wrote. I think you are looking at this accurately. At the end of the day what is the best relational way to represent "how can a carbon atom 'bond' with an oxygen atom based solely on properties of both atoms according to Miles' structures?
Airman. I would argue an atom’s slotlayout diagram, including its possible bonding points is a 3-D configuration mapped onto a 2-D surface for convenience. No 3-D information is lost, each bonding slot is well described, i.e. the SL isn’t exactly a 2-D table.Cr6 wrote. 2-D style tables can sort of do it but graph databases-frame may allow more properties for linking instead of just wide dataframe columns.
Otherwise, agreed. As far as I can tell, when describing atoms or molecules, graph databases are flexible and standard.
Airman. Granted. I expect each atoms’ bonding sites need to be identified by certain specific conditions yet to be identified, i.e. building a bonding matrix and/or connection tables. For starters, I wouldn't expect anything too elegant.Cr6 wrote. It is a question of why or why not can these two atoms link or chain with Miles' structures? Graph databases can allow that a bit better in a query.
Airman. I can certainly see vertices-edges in charge field alignments, especially if we were talking about the atom’s proton configuration – i.e. it’s SL diagram. I must say, in all the graphs I’ve seen thus far, any atom within some molecule is never represented by more than a single vertex which, by itself cannot indicate any charge field alignment. I consider that to be a most important hole in current graph theory, one that the charge field can fill.Cr6 wrote. I see the graph vertexes-edges a lot like the charge field alignments. A lot of graph databases can represent social networks based on properties and edge relationships between members.
I believe high powered graphframe theory/applications used by the universities, pharmaceutical companies and the like were - in part, developed in order for computers to discern molecular details without benefit of any atomic model to start with. I suppose it would be ideal to express MM cf SL atomic structures as an expansion of graph vertices within some commonly used molecular graph format.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Weekly Friday update, continuing my slow review of the Pandas User Guide. During the week Anaconda let me know its latest version was available for download and installation, so I allowed it, then used anaconda to update pandas, numpy, python and several other already out-of-date python packages currently installed in my main working environment, surprised that they total almost 300. The great majority of which I cannot account for – except perhaps as many multiple related version packages or dependencies(??).
Checked to see that pandas.plot() - which imports matplotlib plot(), was now operating properly so I went back to section 15 - Chart visualization. With it’s wide variety of plot diagrams and methods to review, the user guide’s prettiest section. I’m definitely a visual learner.
Currently in section 24 - Scaling to large datasets.
This break can’t last much longer. For all I know, there are likely plenty of alternatives, nothing simple has occurred to me yet. I occasionally have brief glimmers of how what I’m looking at may or may not apply to the project; i.e. a windowing operation may be better suited to calculate and create a bond between two atomic slots compared to groupby()(??).
More later.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
I uploaded a slightly modified MathisPeriodicTable .csv with column header names changed so that works with Jupyter-Databricks:
https://www.mediafire.com/file/jze5jd7qi1sme79/MathisPeriodicTable.csv/file
import pyspark
from pyspark.sql import SparkSession
from pyspark.sql.types import *
import pyspark.sql.functions as F
I'm using the following code for StructTypes - and loading:
schemaPT = StructType([
StructField("UID",IntegerType(),True),
StructField("AtomicSymbol",StringType(),True),
StructField("Element",StringType(),True),
StructField("mOrbitalGroup",StringType(),True),
StructField("mProtons",IntegerType(),True),
StructField("mNeutrons",IntegerType(),True),
StructField("mPeriod",IntegerType(),True),
StructField("SlotOrder",IntegerType(),True),
StructField("AlphaType",IntegerType(),True),
StructField("s1",StringType(),True),
StructField("s2",StringType(),True),
StructField("s3",StringType(),True),
StructField("s4",StringType(),True),
StructField("s5",StringType(),True),
StructField("s6",StringType(),True),
StructField("s7",StringType(),True),
StructField("s8",StringType(),True),
StructField("s9",StringType(),True),
StructField("s10",StringType(),True),
StructField("s11",StringType(),True),
StructField("s12",StringType(),True),
StructField("s13",StringType(),True),
StructField("s14",StringType(),True),
StructField("s15",StringType(),True),
StructField("s16",StringType(),True),
StructField("s17",StringType(),True),
StructField("s18",StringType(),True),
StructField("s19",StringType(),True),
StructField("s20",StringType(),True),
StructField("RemainderAlphaLevel1",IntegerType(),True),
StructField("Rem2",IntegerType(),True),
StructField("Rem3",IntegerType(),True),
StructField("Rem4",IntegerType(),True),
StructField("AlphaTypeRemainder",IntegerType(),True),
StructField("MathisCarouselAlphaTypeFull",IntegerType(),True),
StructField("AtomicNumber",IntegerType(),True),
StructField("OrbitalGroup",StringType(),True),
StructField("PositiveIon",IntegerType(),True),
StructField("NegativeIon",IntegerType(),True),
StructField("Protons",IntegerType(),True),
StructField("Electrons",IntegerType(),True),
StructField("Neutrons",IntegerType(),True),
StructField("PercDiff1stIonization",DoubleType(),True),
StructField("Ions",StringType(),True),
StructField("GroupNumber",IntegerType(),True),
StructField("OldGroupNumber",StringType(),True),
StructField("Period",IntegerType(),True),
StructField("AtomicMassRounded",DoubleType(),True),
StructField("atomic_mass_IUPAC_2005",DoubleType(),True),
StructField("Melting_Point_Celsius",DoubleType(),True),
StructField("Boiling_Point_Celsius",DoubleType(),True),
StructField("State_at_Room_Temp",StringType(),True),
StructField("density_g_mL52",DoubleType(),True),
StructField("electronegativity_Pauling",DoubleType(),True),
StructField("_1st_ionization_potential_eV",DoubleType(),True),
StructField("_2nd_ionization_potential_eV",DoubleType(),True),
StructField("_3rd_ionization_potential_eV",DoubleType(),True),
StructField("_1st_ionization_potential_kJ_mol",IntegerType(),True),
StructField("_2nd_ionization_potential_kJ_mol",IntegerType(),True),
StructField("_3rd_ionization_potential_kJ_mol",IntegerType(),True),
StructField("Electron_Affinity_eV",DoubleType(),True),
StructField("Electron_Affinity_kJ_mol",IntegerType(),True),
StructField("common_oxidation_states",StringType(),True),
StructField("ions_commonly_formed",StringType(),True),
StructField("Predicted_Electron_Configuration",StringType(),True),
StructField("Observed_Electron_Configuration",StringType(),True),
StructField("atomic_radius_",DoubleType(),True),
StructField("ionic_radius_",DoubleType(),True),
StructField("covalent_radius_",DoubleType(),True),
StructField("radius_2n_ion_pm",IntegerType(),True),
StructField("radius_1n_ion_pm",StringType(),True),
StructField("atomic_radius_pm",DoubleType(),True),
StructField("radius_1p_ion_pm",IntegerType(),True),
StructField("radius_2p_ion_pm",DoubleType(),True),
StructField("radius_3p_ion_pm",DoubleType(),True),
StructField("atomic_volume_cm3_mol",DoubleType(),True),
StructField("crystal_structure",StringType(),True),
StructField("CAS_Registry_No",StringType(),True),
StructField("electrical_conductivity_mho_cm",DoubleType(),True),
StructField("specific_heat_J_g_K",DoubleType(),True),
StructField("heat_of_fusion_kJ_mol",DoubleType(),True),
StructField("heat_of_vaporization_kJ_mol",DoubleType(),True),
StructField("thermal_conductivity_W_m_K",DoubleType(),True),
StructField("mg_kg_in_Earths_crust",DoubleType(),True),
StructField("mg_L_in_seawater",DoubleType(),True),
StructField("perc_human_body_mass",DoubleType(),True),
StructField("Notes",StringType(),True),
StructField("source",StringType(),True),
StructField("color",StringType(),True),
StructField("characteristics",StringType(),True),
StructField("structure",StringType(),True),
StructField("hardness_mohs",DoubleType(),True),
StructField("uses",StringType(),True),
StructField("reaction_with_air",StringType(),True),
StructField("reaction_with_water",StringType(),True),
StructField("reaction_with_6M_HCl",StringType(),True),
StructField("reaction_with_15M_HNO3",StringType(),True),
StructField("reaction_with_6M_NaOH",StringType(),True),
StructField("hydride",StringType(),True),
StructField("oxide",StringType(),True),
StructField("chloride",StringType(),True),
StructField("polarizability_A3",DoubleType(),True),
StructField("heat_atomization_kJ_mol",IntegerType(),True),
StructField("year_discovered",IntegerType(),True),
StructField("rel_abund_solar_system_log",DoubleType(),True),
StructField("abundance_earths_crust_log",DoubleType(),True),
StructField("cost_pure_100g",DoubleType(),True),
StructField("cost_bulk_100g",DoubleType(),True),
StructField("toxic",StringType(),True),
StructField("carcinogenic",StringType(),True),
StructField("PeriodicTableID",IntegerType(),True),
StructField("Name",StringType(),True),
StructField("UID3",IntegerType(),True),
StructField("Symbol",StringType(),True),
StructField("BP_K",DoubleType(),True),
StructField("MP_K",DoubleType(),True),
StructField("Density",DoubleType(),True),
StructField("AtomicWeight",DoubleType(),True),
StructField("MostStableOxidationState",IntegerType(),True),
StructField("CovalentRadius",DoubleType(),True),
StructField("AtomicNumberRadiusAngstroms",DoubleType(),True),
StructField("FirstIP",DoubleType(),True),
StructField("SpecificHeatCapacity",DoubleType(),True),
StructField("ThermalConductivity",DoubleType(),True),
StructField("ElectricalConductivity",DoubleType(),True),
StructField("HeatOfFusion",DoubleType(),True),
StructField("HeatOfVaporization",DoubleType(),True),
StructField("ElectroNegativity",DoubleType(),True),
StructField("YearsSinceDiscovery",IntegerType(),True),
StructField("Family",StringType(),True),
StructField("Group",IntegerType(),True),
StructField("orbital",StringType(),True),
StructField("MetalOrNonMetal",StringType(),True),
StructField("_2ndmoststableoxidationstate",IntegerType(),True),
StructField("_3rdmoststableoxidationstate",IntegerType(),True),
StructField("_4thmoststableoxidationstate",IntegerType(),True),
StructField("mg_kg_crust",DoubleType(),True),
StructField("mg_L_seawater",DoubleType(),True),
StructField("UID2",DoubleType(),True),
StructField("length_of_name",IntegerType(),True),
StructField("Series",StringType(),True),
StructField("StateofMatter",StringType(),True),
StructField("Atomic_Radius_A",DoubleType(),True),
StructField("Boiling_point_oC",DoubleType(),True),
StructField("Melting_Freezing_Point_oC",DoubleType(),True),
StructField("Density_g_mL145",DoubleType(),True),
StructField("PeriodNumber",IntegerType(),True),
StructField("PeriodTypeDescription",StringType(),True),
StructField("LithosphereAbundance",IntegerType(),True),
StructField("RelativeProportion_ppm",DoubleType(),True),
StructField("CrustalAbundance1_ppm",DoubleType(),True),
StructField("CrustalAbundance2_ppm",IntegerType(),True),
StructField("CrustalAbundance3_ppm",IntegerType(),True),
StructField("ProductionTonnes",IntegerType(),True),
StructField("a1s",StringType(),True),
StructField("a2s",StringType(),True),
StructField("a2p",StringType(),True),
StructField("a3s",StringType(),True),
StructField("a3p",StringType(),True),
StructField("a3d",StringType(),True),
StructField("a4s",StringType(),True),
StructField("a4p",StringType(),True),
StructField("a4d",StringType(),True),
StructField("a4f",StringType(),True),
StructField("a5s",StringType(),True),
StructField("a5p",StringType(),True),
StructField("a5d",StringType(),True),
StructField("a5f",StringType(),True),
StructField("a5g",StringType(),True),
StructField("a6s",StringType(),True),
StructField("a6p",StringType(),True),
StructField("a6d",StringType(),True),
StructField("a6f",StringType(),True),
StructField("a7s",StringType(),True),
StructField("a7p",StringType(),True),
StructField("a7d",StringType(),True),
StructField("a8s",StringType(),True),
StructField("a8p",StringType(),True),
StructField("a9s",StringType(),True),
StructField("a9p",StringType(),True)])
# File location and type
file_location = "/MathisPeriodicTable.csv"
file_type = "csv"
# CSV options
infer_schema = "false"
first_row_is_header = "true"
delimiter = ","
df = spark.read.format(file_type).schema(schemaPT).options(header=first_row_is_header, inferSchema=infer_schema, delimiter=delimiter).load(file_location)
df.printSchema
df.display()
permanent_table_name = "MathisPeriodicPermTable"
df.write.format("parquet").saveAsTable(permanent_table_name)

Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Downloaded and quickly reviewed “MathisPeriodicTable.csv”. Plenty of data. 1691 rows by 180 columns. The image shows a list from Hydrogen (H,1) through Hafnium (Hf,72), although the data itself appears sufficient to build all the atoms through Ununoctium (Uuo,118).
One problem prevents me from uploading the file to github. The data for Lithium (Li,3) is missing.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Let me look into it. It may be an older copy.
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Ok, made it through the pandas User Guide. The final section - Cookbook, didn’t contain many examples, although it does contain plenty of subject matter links I’ve yet to visit. Looking for a more concrete example I started working a RMS Titanic passenger list data tutorial, using “titanic.csv”.
https://pandas.pydata.org/docs/getting_started/intro_tutorials/03_subset_data.html#min-tut-03-subset
I also made it through Numpy : the absolute basics for beginners, and am now in Numpy fundamentals.
https://numpy.org/doc/stable/user/basics.indexing.html
Before considering any molecules, I believe I’m just about ready to resume where I left off. Creating Version 2 of Atom Builder, wherein the approximately 2,000 code lines making up the 19 i-loops generating the desired atom will be replaced with a dataFrame containing each atom’s slotlayout values taken from Cr6-Elements.csv, or MathisPeriodicTable.csv.
As is, to view a new atom, one must go back to the initial periodic table or the control panel’s element dropdown list in order to change the selection, then rerun most of the Atom Builder notebook to view the output. Changing to a dataframe should enable more efficient code, such as allowing a user to select a new atom from a new control tab within the final tab enclosure itself – would be a big improvement. Plus, last Saturday I noticed an Atom Builder python warning associated with the atomic label message dimension being out-of-range.
With an eye toward molecules I must consider how best to access and modify the current csv table.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Status. Again, sorry for the delay, I hope I haven’t caused any distress. I’m working at it all right, in fact I like working at it and never fail to spend at least a couple of hours at it every day. My only problem is, I’ve yet to make much - if any, tangible progress. No surprise, learning is taking me longer than I’d hoped.
I have no problem switching to MathisPeriodicTable.csv whenever you like. It is more complicated, for example, the number of rows for each element is equal to that element’s (1-118) number of proton occupied slots (1-19). Its dataFrame (all elements including Li) should be 1693 rows by 180 columns. MathisPeriodicTable’s main complexity involves the fact that all the csv’s data should be made available to the notebook user for review, necessitating new and likely extensive user interfaces, like you showed in your 23 January, Jupyter-Databricks PercDiff1stIonization plot above – just one of the 180 dataFrame columns. On the other hand, delaying a bit is also, not a problem. I repeat, I have no objection to switching to MathisPeriodicTable either sooner or later. Your directions are needed.
As previously mentioned, I’ve modified Cr6-Elements.csv which currently contains 19 rows for element, H,1 thrugh Th,90. The dataFrame contains 1710 rows × 23 columns. The 23 column headers are shown in Out [7] below.
My present efforts are concerned with reviewing the data types of each of the columns I’ll need to access.
- Code:
In [5]: filename = '../data/Cr6-Elements.csv'
In [6]: df = pd.read_csv(filename)
In [7]: df.dtypes
Out [7]:
AtomicNumber int64
AtomicSymbol object
Element object
AtomicTuple object
OrbitalGroup object
Protons int64
Neutrons int64
GroupNumber float64
Period int64
SlotNumber int64
SlotP int64
SlotN int64
SlotOrien object
AlphaType object
SlotX int64
SlotY int64
SlotZ int64
CanBind object
SlotLayout object
SL1 object
SL2 object
SL3 object
SL4 object
dtype: object
Any given column should contain only a single correct data type, that is currently not the case. Strings, such as “AtomicSymbol”, “Element” and “AtomicTuple” (actually a tuple) etc. are identified as “object” which may also include unnecessary NaN values. I removed the first 38 NaN’s from the top of the “OrbitalGroup” column, the first “OrbitalGroup” value occurs at Li which is [He] in row 39, replacing each blank with a dummy value [Top], or maybe [Zero] would make more sense. There are also NaN values in the “GroupNumber” column. There are also many NaN values in the final 5 columns, “SlotLayout” through “SL4” which I probably will not need to access.
Another item, I don’t believe the “AtomicNumber” type should be an integer, or int64 dtype. Each 1-90 numbers are unique, corresponding to the 1-90 elements, 90 different “category” values which pandas treats differently than integer types. The values in the “AtomicSymbol” and “Element” columns should also be considered as 90 unique categories within each column rather than string objects. Likewise, “SlotOrien” is either H or V for horizontal or vertical proton emission plane orientation, just two possible categories. The same is true for the “CanBind” column which contains either yes or no - Y or N category values.
How best to make those changes? Going back over “Data types”, “I/O Tools” and “Category” user guide sections again.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Maybe a little progress.
For starters I’m still making Cr6-Elements.csv file changes:
1. "OrbitalGroup". I suppose this column identifies each element’s core configuration, excluding Hydrogen and Helium which do not have a core to build upon, (i.e, the third proton of Lithium builds on a He core), with possible values: [He],[Ne],[Ar],[Kr],[Xe], and [Rn]. Along that line, Miles has mentioned that the alpha unit, a.k.a. He, should be considered the first stable element; H, a single, lone proton cannot pair with a neutron for long since the neutron is too exposed on its side opposite the proton. Last time I added a new value [Top] to fill the first 38 blank (2x19) entries. This time I changed ‘Top’ to ‘Zero’ and removed 1710 square brackets from each row’s entry, i.e. from [He] to He.
2. “GroupNumber”. The Periodic table columns, 1-18, left-right sides. Contained blank entries for elements 57-71, the Lanthanides; filled those blanks with a new dummy value, ’19’. Also filled the blanks for elements 89-90, the Actinides, our largest atoms, with new dummy value ‘20’,
3. "SlotOrien". Slot proton emission orientation, values H or V. Filled two blanks, in row 720 Strontium slot 16 with ‘H’, and in row 990 Iodine slot 1 with ‘V’, according to their slot diagrams, columns 18-22.
4. "CanBind". Values ‘Y’ or ‘No’. Zero values in rows 135 Oxygen slot 1 and 154 Fluorine slot 1 changed to “Y”. I realize that slot 1 cannot bind with anything and so I’ll likely change both to “N” at some point in the future.
5. "AlphaType". Stack proton count (1-6) for that row’s atomic slot (1-19). Verified two corrections using Atom Builder. Yttrium, row 729, slot 6, changed a ‘d’ to “4”; and changed a ‘d’ in row 1259, Holmium, slot 4, to a proton count of ‘5’.
Note, if I start making real progress I’ll continue making Cr6-Elements.csv changes, such as when adding slot coordinates.
Next, I made changes to the pandas.read_csv() default data type values, included in the code below. Last time I failed to mention that the data types shown – “objects”, “int64” and “float64” were created automatically by pandas. The default values are usually good enough to work with, but one should always make sure. I thought I would change the "AtomicNumber” from “int64” and several other “object” data type columns to a “category” type instead, and perhaps a few other changes.
Went back to the user guide and made a few searches. I must mention a really nice set of postings from a python/pandas blog, Practical Business Python written by Chris Moffitt *. Kinda scary downloading and running a 600 Mega byte historical medical data file just to prove a few points, but it was no problem at all. Object data can actually contain multiple different types including numbers and strings. Chris makes it clear one should use “category” type data when dealing with very large files that can cause memory limit problems because the “category” type uses maybe a sixth to a tenth the memory required by an “object” type data. Category data can be used to create either logical or lexical order to an object series when converting to a category type, which can greatly increase code performance. On the other hand, one must be extremely careful when working with category types, there are many easy ways to screw-up; best to avoid, unless needed. Message received and understood. At present, other than listing categorical order, I don’t see any real need to convert any data columns into a category type.
Here are my few data type code changes along with comments and the resulting df. I didn't show it, but last time, "Memory usage: 284.0+ KB". Now "memory usage: 217.2+ KB".
- Code:
In [1]: import pandas as pd
In [2]: import numpy as np
In [3]: filename = '../data/Cr6-Elements.csv'
In [4]: df = pd.read_csv(filename)
In [5]: # Make "SlotOrien" and "CanBind" boolean.
df["SlotOrien"] = df["SlotOrien"].map({"H":True, "V":False})
In [6]: df["CanBind"] = np.where(df["CanBind"] == "Y", True, False)
In [7]: # Make “SlotX”, “SlotY” and “SlotZ”, a slot’s x y and z coordinates float values.
df["SlotX"] = df["SlotX"].astype("float")
In [8]: df["SlotY"] = df["SlotY"].astype("float")
In [9]: df["SlotZ"] = df["SlotZ"].astype("float")
In [10]: df = df.iloc[:,:18]
# “SlotLayout”-“Slot4”, cols (18-22) aren’t removed,
# they are simply not read by the working dataFrame df.
In [11]: df.info()
Out [11]:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1710 entries, 0 to 1709
Data columns (total 18 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 AtomicNumber 1710 non-null int64
1 AtomicSymbol 1710 non-null object
2 Element 1710 non-null object
3 AtomicTuple 1710 non-null object
4 OrbitalGroup 1710 non-null object
5 Protons 1710 non-null int64
6 Neutrons 1710 non-null int64
7 GroupNumber 1710 non-null int64
8 Period 1710 non-null int64
9 SlotNumber 1710 non-null int64
10 SlotP 1710 non-null int64
11 SlotN 1710 non-null int64
12 SlotOrien 1710 non-null bool
13 AlphaType 1710 non-null int64
14 SlotX 1710 non-null float64
15 SlotY 1710 non-null float64
16 SlotZ 1710 non-null float64
17 CanBind 1710 non-null bool
dtypes: bool(2), float64(3), int64(9), object(4)
memory usage: 217.2+ KB
*
Practical Business Python
https://pbpython.com/
Overview of Pandas Data Types
https://pbpython.com/pandas_dtypes.html
Using The Pandas Category Data Type
https://pbpython.com/pandas_dtypes_cat.html
Tips for Selecting Columns in a DataFrame
https://pbpython.com/selecting-columns.html
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Significant milestone progress. The ‘elements’ tuple series is now entirely derived from Cr6-Elements. Re-selecting the desired atom is working properly.
- Code:
In[12]: # Recreate 'elements', the tuple series used by the
# program to identify or reselect a desired atom from
# the gui dropdown or periodic table.
elements_x = df['AtomicTuple'].drop_duplicates()
elements_x
Out[12]:
0 (hydrogen, 1)
19 (helium, 2)
38 (lithium, 3)
57 (beryllium, 4)
76 (boron, 5)
...
1615 (radon, 86)
1634 (francium, 87)
1653 (radium, 88)
1672 (actinium, 89)
1691 (thorium, 90)
Name: AtomicTuple, Length: 90, dtype: object
In[13]: # elements_x's row index increments by 19's, 0 to 1709,
# 19x expanded. The Series.array method renumbers the
# index back to the desired 0-89.
elements = elements_x.array
- Code:
df['AtomicTuple'] = list(zip(df.Element, df.AtomicNumber))
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
- Code:
In[1]:
Molecules
Molecules will render a 3D charge field model of molecular
matter from atoms selected by the user.
That's the plan anyway. At first I'll try making atoms.
Molecules is Version 2 of Atom Builder (AB), replacing
AB's 19 i-loop slots with a 19 slot csv data file. The
expected faster renderings and efficiencies gained
should allow us to build multiple atoms in molecular
configurations.
I might end up using the json data file instead. I think
I'll try reusing most of AB's code, such as the periodic
table. This time around we'll use Cr6's original Slotlayout
(SL) diagram.
Molecules is intended to introduce Charge field theory
- see the notebook's end cell's source references.
In[2]:
from pythreejs import *
from IPython.display import display
from ipywidgets import HBox, VBox, Button, ButtonStyle, GridspecLayout
import ipywidgets as widgets
from math import sqrt, tan
from random import random , randint , seed
In[3]:
# A few values used
phi = (1+sqrt(5))/2 # calculating phi-based-pi, piG.
piG = 4/sqrt(phi) # piG is used instead of pi.
pRadius = 0.05 # proton radius, used by all atomic distances
caroSecs = 5.0 # Problem Here # time in secs for the carousel to make one rotation
In[4]:
# Sizing the view, camera and adding lights
view_width = 600
view_height = 400
camera = PerspectiveCamera(position=[5, 0, 0], up=[0, 0, 1], aspect=view_width/view_height)
#camera = PerspectiveCamera(position=[5, 0, 0], up=[1, 0, 0], aspect=view_width/view_height)
camera = PerspectiveCamera(position=[5, 0, 0], up=[0, 1, 0], aspect=view_width/view_height)
key_light = DirectionalLight(position=[0, 5, 5])
ambient_light = AmbientLight()
In[5]:
import pandas as pd
import numpy as np
#pd.set_option('max_columns', 4, 'max_rows', 10, 'max_colwidth', 12)
filename = '../data/Cr6-Elements.csv'
df_raw = pd.read_csv(filename)
# Uncomment df_raw or df_raw.info() to view
# df_raw
# df_raw.info()
In[6]: df = df_raw.copy()
# Make changes to the dataFrame created by pd.read_csv(filename)'s
# method defaults. The data type constrains how you may work with
# the data.
# Make "SlotOrien" and "CanBind" boolean.
df["CanBind"] = np.where(df["CanBind"] == "Y", True, False)
df["SlotOrien"] = df["SlotOrien"].map({"H":True, "V":False})
# Slot coordinates need to be float
df["SlotX"] = df["SlotX"].astype("float")
df["SlotY"] = df["SlotY"].astype("float")
df["SlotZ"] = df["SlotZ"].astype("float")
# Do not include the SlotLayout columns.
df = df.iloc[:,:19]
# Uncomment df or df.info() to view
# df
# df.info()
In[7]:
"""
'elements'is a list of tuples used by the program to
identify or re-select a desired atom from the gui
dropdown or periodic table. Also used as the optional
3D atom's atomic label. 'elements' is taken from the
df['AtomicTuple'] column.
Starting with 1710 rows. After drop_duplicates, the
'AtomicTuple' row index increments by 19's, 0 to 1709,
19x expanded. The Series.array method renumbers the
index back to the desired 0-89. The two steps are
chained.
Next, atomicTups has been read as a list of strings.
Those strings are converted into tuples by using
the map and eval methods.
I found the conversion by searching "How to convert
a list of strings into tuples in python?". It works.
"""
atomicTups = df['AtomicTuple'].drop_duplicates().array
elements = list(map(eval, atomicTups))
In[8]: # The periodic table's 90 buttons.
# Each button includes: atomic symbol, color, layout width and layout height
# 10 colors/element types:alkali metal, alkaline earth, transition metal,basic
# metal, metalloid, nonmetal, halogen, noble gas, lanthanide, actinide. Plus an
# unused 11th color - blueviolet.# Stopping at violet causes an out of limit error(??)
clrs = ['tomato', 'orange','yellow','palegreen','springgreen','skyblue','powderblue',
'cyan','thistle','violet','blueviolet' ]
# The colors for each of the atoms, 1-90.
clist = [clrs[5],clrs[7],clrs[0],clrs[1],clrs[4],clrs[5],clrs[5],clrs[5],clrs[6],clrs[7],
clrs[0],clrs[1],clrs[3],clrs[4],clrs[5],clrs[5],clrs[6],clrs[7],clrs[0],clrs[1],
clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],
clrs[3],clrs[4],clrs[4],clrs[5],clrs[6],clrs[7],clrs[0],clrs[1],clrs[2],clrs[2],
clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[3],clrs[3],
clrs[4],clrs[4],clrs[6],clrs[7],clrs[0],clrs[1],clrs[8],clrs[8],clrs[8],clrs[8],
clrs[8],clrs[8],clrs[8],clrs[8],clrs[8],clrs[8],clrs[8],clrs[8],clrs[8],clrs[8],
clrs[8],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],clrs[2],
clrs[3],clrs[3],clrs[3],clrs[4],clrs[6],clrs[7],clrs[0],clrs[1],clrs[9],clrs[9]]
# The atomic symbols for the Periodic table. Handled like elements.
dlist = df['AtomicSymbol'].drop_duplicates().array
# Defining the buttons by adding the atomic symbols and colors to the buttons
a1 = Button(description=dlist[0], style=dict(button_color=clist[0]))
a2 = Button(description=dlist[1], style=dict(button_color=clist[1]))
a3 = Button(description=dlist[2], style=dict(button_color=clist[2]))
a4 = Button(description=dlist[3], style=dict(button_color=clist[3]))
a5 = Button(description=dlist[4], style=dict(button_color=clist[4]))
a6 = Button(description=dlist[5], style=dict(button_color=clist[5]))
a7 = Button(description=dlist[6], style=dict(button_color=clist[6]))
a8 = Button(description=dlist[7], style=dict(button_color=clist[7]))
a9 = Button(description=dlist[8], style=dict(button_color=clist[8]))
a10 = Button(description=dlist[9], style=dict(button_color=clist[9]))
a11 = Button(description=dlist[10], style=dict(button_color=clist[10]))
a12 = Button(description=dlist[11], style=dict(button_color=clist[11]))
a13 = Button(description=dlist[12], style=dict(button_color=clist[12]))
a14 = Button(description=dlist[13], style=dict(button_color=clist[13]))
a15 = Button(description=dlist[14], style=dict(button_color=clist[15]))
a16 = Button(description=dlist[15], style=dict(button_color=clist[15]))
a17 = Button(description=dlist[16], style=dict(button_color=clist[16]))
a18 = Button(description=dlist[17], style=dict(button_color=clist[17]))
a19 = Button(description=dlist[18], style=dict(button_color=clist[18]))
a20 = Button(description=dlist[19], style=dict(button_color=clist[19]))
a21 = Button(description=dlist[20], style=dict(button_color=clist[20]))
a22 = Button(description=dlist[21], style=dict(button_color=clist[22]))
a23 = Button(description=dlist[22], style=dict(button_color=clist[22]))
a24 = Button(description=dlist[23], style=dict(button_color=clist[23]))
a25 = Button(description=dlist[24], style=dict(button_color=clist[24]))
a26 = Button(description=dlist[25], style=dict(button_color=clist[25]))
a27 = Button(description=dlist[26], style=dict(button_color=clist[26]))
a28 = Button(description=dlist[27], style=dict(button_color=clist[27]))
a29 = Button(description=dlist[28], style=dict(button_color=clist[28]))
a30 = Button(description=dlist[29], style=dict(button_color=clist[29]))
a31 = Button(description=dlist[30], style=dict(button_color=clist[30]))
a32 = Button(description=dlist[31], style=dict(button_color=clist[31]))
a33 = Button(description=dlist[32], style=dict(button_color=clist[32]))
a34 = Button(description=dlist[33], style=dict(button_color=clist[33]))
a35 = Button(description=dlist[34], style=dict(button_color=clist[34]))
a36 = Button(description=dlist[35], style=dict(button_color=clist[35]))
a37 = Button(description=dlist[36], style=dict(button_color=clist[36]))
a38 = Button(description=dlist[37], style=dict(button_color=clist[37]))
a39 = Button(description=dlist[38], style=dict(button_color=clist[38]))
a40 = Button(description=dlist[39], style=dict(button_color=clist[39]))
a41 = Button(description=dlist[40], style=dict(button_color=clist[40]))
a42 = Button(description=dlist[41], style=dict(button_color=clist[41]))
a43 = Button(description=dlist[42], style=dict(button_color=clist[42]))
a44 = Button(description=dlist[43], style=dict(button_color=clist[43]))
a45 = Button(description=dlist[44], style=dict(button_color=clist[44]))
a46 = Button(description=dlist[45], style=dict(button_color=clist[45]))
a47 = Button(description=dlist[46], style=dict(button_color=clist[46]))
a48 = Button(description=dlist[47], style=dict(button_color=clist[47]))
a49 = Button(description=dlist[48], style=dict(button_color=clist[48]))
a50 = Button(description=dlist[49], style=dict(button_color=clist[49]))
a51 = Button(description=dlist[50], style=dict(button_color=clist[50]))
a52 = Button(description=dlist[51], style=dict(button_color=clist[51]))
a53 = Button(description=dlist[52], style=dict(button_color=clist[52]))
a54 = Button(description=dlist[53], style=dict(button_color=clist[53]))
a55 = Button(description=dlist[54], style=dict(button_color=clist[54]))
a56 = Button(description=dlist[55], style=dict(button_color=clist[55]))
a57 = Button(description=dlist[56], style=dict(button_color=clist[56]))
a58 = Button(description=dlist[57], style=dict(button_color=clist[57]))
a59 = Button(description=dlist[58], style=dict(button_color=clist[58]))
a60 = Button(description=dlist[59], style=dict(button_color=clist[59]))
a61 = Button(description=dlist[60], style=dict(button_color=clist[60]))
a62 = Button(description=dlist[61], style=dict(button_color=clist[61]))
a63 = Button(description=dlist[62], style=dict(button_color=clist[62]))
a64 = Button(description=dlist[63], style=dict(button_color=clist[63]))
a65 = Button(description=dlist[64], style=dict(button_color=clist[64]))
a66 = Button(description=dlist[65], style=dict(button_color=clist[65]))
a67 = Button(description=dlist[66], style=dict(button_color=clist[66]))
a68 = Button(description=dlist[67], style=dict(button_color=clist[67]))
a69 = Button(description=dlist[68], style=dict(button_color=clist[68]))
a70 = Button(description=dlist[69], style=dict(button_color=clist[69]))
a71 = Button(description=dlist[70], style=dict(button_color=clist[70]))
a72 = Button(description=dlist[71], style=dict(button_color=clist[71]))
a73 = Button(description=dlist[72], style=dict(button_color=clist[72]))
a74 = Button(description=dlist[73], style=dict(button_color=clist[73]))
a75 = Button(description=dlist[74], style=dict(button_color=clist[74]))
a76 = Button(description=dlist[75], style=dict(button_color=clist[75]))
a77 = Button(description=dlist[76], style=dict(button_color=clist[76]))
a78 = Button(description=dlist[77], style=dict(button_color=clist[77]))
a79 = Button(description=dlist[78], style=dict(button_color=clist[78]))
a80 = Button(description=dlist[79], style=dict(button_color=clist[79]))
a81 = Button(description=dlist[80], style=dict(button_color=clist[80]))
a82 = Button(description=dlist[81], style=dict(button_color=clist[81]))
a83 = Button(description=dlist[82], style=dict(button_color=clist[82]))
a84 = Button(description=dlist[83], style=dict(button_color=clist[83]))
a85 = Button(description=dlist[84], style=dict(button_color=clist[84]))
a86 = Button(description=dlist[85], style=dict(button_color=clist[85]))
a87 = Button(description=dlist[86], style=dict(button_color=clist[86]))
a88 = Button(description=dlist[87], style=dict(button_color=clist[87]))
a89 = Button(description=dlist[88], style=dict(button_color=clist[88]))
a90 = Button(description=dlist[89], style=dict(button_color=clist[89]))
# Creating the list of buttons
abttns = [a1,a2,a3,a4,a5,a6,a7,a8,a9,a10,a11,a12,a13,a14,a15,a16,a17,a18,a19,
a20,a21,a22,a23,a24,a25,a26,a27,a28,a29,a30,a31,a32,a33,a34,a35,a36,a37,a38,a39,
a40,a41,a42,a43,a44,a45,a46,a47,a48,a49,a50,a51,a52,a53,a54,a55,a56,a57,a58,a59,
a60,a61,a62,a63,a64,a65,a66,a67,a68,a69,a70,a71,a72,a73,a74,a75,a76,a77,a78,a79,
a80,a81,a82,a83,a84,a85,a86,a87,a88,a89,a90]
# Use the button list in a loop to add each button's border, layout width and height.
i=0
while i < 90 :
abttns[i].layout = widgets.Layout(border = '1px solid black')
abttns[i].layout.width = '100%'
abttns[i].layout.height = '100%' #'40px'
i += 1
# Layout the 90 buttons in periodic table form
grid = GridspecLayout(10, 18)
grid[0,0] = abttns[0]
grid[0,17] = abttns[1]
grid[1,0] = abttns[2]
grid[1,1] = abttns[3]
p=12
while p < 18:
grid[1,p] = abttns[p-8]
p += 1
grid[2,0] = abttns[10]
grid[2,1] = abttns[11]
n=12
while n < 18:
grid[2,n] = abttns[n]
n += 1
m=0
while m < 18:
grid[3,m] = abttns[18+m]
m += 1
k=0
while k < 18:
grid[4,k] = abttns[36+k]
k += 1
grid[5,0] = abttns[54]
grid[5,1] = abttns[55]
# grid[5,2] = the Lanthanide series hole
i=3
while i < 18:
grid[5,i] = abttns[68+i]
i += 1
grid[6,0] = abttns[86]
grid[6,1] = abttns[87]
# grid[6,2] = the Actinide series hole
j=2
while j < 17:
# Lanthanide series
grid[7,j] = abttns[54+j]
j += 1
#Actinide series
grid[8,2] = abttns[88]
grid[8,3] = abttns[89]
# The periodic table button functions
output = widgets.Output()
#display(output)
@output.capture()
def on_a1_clicked(a1): selectAtom.value = 1
def on_a2_clicked(a2): selectAtom.value = 2
def on_a3_clicked(a3): selectAtom.value = 3
def on_a4_clicked(a4): selectAtom.value = 4
def on_a5_clicked(a5): selectAtom.value = 5
def on_a6_clicked(a6): selectAtom.value = 6
def on_a7_clicked(a7): selectAtom.value = 7
def on_a8_clicked(a8): selectAtom.value = 8
def on_a9_clicked(a9): selectAtom.value = 9
def on_a10_clicked(a10): selectAtom.value = 10
def on_a11_clicked(a11): selectAtom.value = 11
def on_a12_clicked(a12): selectAtom.value = 12
def on_a13_clicked(a13): selectAtom.value = 13
def on_a14_clicked(a14): selectAtom.value = 14
def on_a15_clicked(a15): selectAtom.value = 15
def on_a16_clicked(a16): selectAtom.value = 16
def on_a17_clicked(a17): selectAtom.value = 17
def on_a18_clicked(a18): selectAtom.value = 18
def on_a19_clicked(a19): selectAtom.value = 19
def on_a20_clicked(a20): selectAtom.value = 20
def on_a21_clicked(a21): selectAtom.value = 21
def on_a22_clicked(a22): selectAtom.value = 22
def on_a23_clicked(a23): selectAtom.value = 23
def on_a24_clicked(a24): selectAtom.value = 24
def on_a25_clicked(a25): selectAtom.value = 25
def on_a26_clicked(a26): selectAtom.value = 26
def on_a27_clicked(a27): selectAtom.value = 27
def on_a28_clicked(a28): selectAtom.value = 28
def on_a29_clicked(a29): selectAtom.value = 29
def on_a30_clicked(a30): selectAtom.value = 30
def on_a31_clicked(a31): selectAtom.value = 31
def on_a32_clicked(a32): selectAtom.value = 32
def on_a33_clicked(a33): selectAtom.value = 33
def on_a34_clicked(a34): selectAtom.value = 34
def on_a35_clicked(a35): selectAtom.value = 35
def on_a36_clicked(a36): selectAtom.value = 36
def on_a37_clicked(a37): selectAtom.value = 37
def on_a38_clicked(a38): selectAtom.value = 38
def on_a39_clicked(a39): selectAtom.value = 39
def on_a40_clicked(a40): selectAtom.value = 40
def on_a41_clicked(a41): selectAtom.value = 41
def on_a42_clicked(a42): selectAtom.value = 42
def on_a43_clicked(a43): selectAtom.value = 43
def on_a44_clicked(a44): selectAtom.value = 44
def on_a45_clicked(a45): selectAtom.value = 45
def on_a46_clicked(a46): selectAtom.value = 46
def on_a47_clicked(a47): selectAtom.value = 47
def on_a48_clicked(a48): selectAtom.value = 48
def on_a49_clicked(a49): selectAtom.value = 49
def on_a50_clicked(a50): selectAtom.value = 50
def on_a51_clicked(a51): selectAtom.value = 51
def on_a52_clicked(a52): selectAtom.value = 52
def on_a53_clicked(a53): selectAtom.value = 53
def on_a54_clicked(a54): selectAtom.value = 54
def on_a55_clicked(a55): selectAtom.value = 55
def on_a56_clicked(a56): selectAtom.value = 56
def on_a57_clicked(a57): selectAtom.value = 57
def on_a58_clicked(a58): selectAtom.value = 58
def on_a59_clicked(a59): selectAtom.value = 59
def on_a60_clicked(a59): selectAtom.value = 60
def on_a61_clicked(a61): selectAtom.value = 61
def on_a62_clicked(a62): selectAtom.value = 62
def on_a63_clicked(a63): selectAtom.value = 63
def on_a64_clicked(a64): selectAtom.value = 64
def on_a65_clicked(a65): selectAtom.value = 65
def on_a66_clicked(a66): selectAtom.value = 66
def on_a67_clicked(a67): selectAtom.value = 67
def on_a68_clicked(a68): selectAtom.value = 68
def on_a69_clicked(a69): selectAtom.value = 69
def on_a70_clicked(a70): selectAtom.value = 70
def on_a71_clicked(a71): selectAtom.value = 71
def on_a72_clicked(a72): selectAtom.value = 72
def on_a73_clicked(a73): selectAtom.value = 73
def on_a74_clicked(a74): selectAtom.value = 74
def on_a75_clicked(a75): selectAtom.value = 75
def on_a76_clicked(a76): selectAtom.value = 76
def on_a77_clicked(a77): selectAtom.value = 77
def on_a78_clicked(a78): selectAtom.value = 78
def on_a79_clicked(a79): selectAtom.value = 79
def on_a80_clicked(a80): selectAtom.value = 80
def on_a81_clicked(a81): selectAtom.value = 81
def on_a82_clicked(a82): selectAtom.value = 82
def on_a83_clicked(a83): selectAtom.value = 83
def on_a84_clicked(a84): selectAtom.value = 84
def on_a85_clicked(a85): selectAtom.value = 85
def on_a86_clicked(a86): selectAtom.value = 86
def on_a87_clicked(a87): selectAtom.value = 87
def on_a88_clicked(a88): selectAtom.value = 88
def on_a89_clicked(a89): selectAtom.value = 89
def on_a90_clicked(a90): selectAtom.value = 90
a1.on_click(on_a1_clicked)
a2.on_click(on_a2_clicked)
a3.on_click(on_a3_clicked)
a4.on_click(on_a4_clicked)
a5.on_click(on_a5_clicked)
a6.on_click(on_a6_clicked)
a7.on_click(on_a7_clicked)
a8.on_click(on_a8_clicked)
a9.on_click(on_a9_clicked)
a10.on_click(on_a10_clicked)
a11.on_click(on_a11_clicked)
a12.on_click(on_a12_clicked)
a13.on_click(on_a13_clicked)
a14.on_click(on_a14_clicked)
a15.on_click(on_a15_clicked)
a16.on_click(on_a16_clicked)
a17.on_click(on_a17_clicked)
a18.on_click(on_a18_clicked)
a19.on_click(on_a19_clicked)
a20.on_click(on_a20_clicked)
a21.on_click(on_a21_clicked)
a22.on_click(on_a22_clicked)
a23.on_click(on_a23_clicked)
a24.on_click(on_a24_clicked)
a25.on_click(on_a25_clicked)
a26.on_click(on_a26_clicked)
a27.on_click(on_a27_clicked)
a28.on_click(on_a28_clicked)
a29.on_click(on_a29_clicked)
a30.on_click(on_a30_clicked)
a31.on_click(on_a31_clicked)
a32.on_click(on_a32_clicked)
a33.on_click(on_a33_clicked)
a34.on_click(on_a34_clicked)
a35.on_click(on_a35_clicked)
a36.on_click(on_a36_clicked)
a37.on_click(on_a37_clicked)
a38.on_click(on_a38_clicked)
a39.on_click(on_a39_clicked)
a40.on_click(on_a40_clicked)
a41.on_click(on_a41_clicked)
a42.on_click(on_a42_clicked)
a43.on_click(on_a43_clicked)
a44.on_click(on_a44_clicked)
a45.on_click(on_a45_clicked)
a46.on_click(on_a46_clicked)
a47.on_click(on_a47_clicked)
a48.on_click(on_a48_clicked)
a49.on_click(on_a49_clicked)
a50.on_click(on_a50_clicked)
a51.on_click(on_a51_clicked)
a52.on_click(on_a52_clicked)
a53.on_click(on_a53_clicked)
a54.on_click(on_a54_clicked)
a55.on_click(on_a55_clicked)
a56.on_click(on_a56_clicked)
a57.on_click(on_a57_clicked)
a58.on_click(on_a58_clicked)
a59.on_click(on_a59_clicked)
a60.on_click(on_a60_clicked)
a61.on_click(on_a61_clicked)
a62.on_click(on_a62_clicked)
a63.on_click(on_a63_clicked)
a64.on_click(on_a64_clicked)
a65.on_click(on_a65_clicked)
a66.on_click(on_a66_clicked)
a67.on_click(on_a67_clicked)
a68.on_click(on_a68_clicked)
a69.on_click(on_a69_clicked)
a70.on_click(on_a70_clicked)
a71.on_click(on_a71_clicked)
a72.on_click(on_a72_clicked)
a73.on_click(on_a73_clicked)
a74.on_click(on_a74_clicked)
a75.on_click(on_a75_clicked)
a76.on_click(on_a76_clicked)
a77.on_click(on_a77_clicked)
a78.on_click(on_a78_clicked)
a79.on_click(on_a79_clicked)
a80.on_click(on_a80_clicked)
a81.on_click(on_a81_clicked)
a82.on_click(on_a82_clicked)
a83.on_click(on_a83_clicked)
a84.on_click(on_a84_clicked)
a85.on_click(on_a85_clicked)
a86.on_click(on_a86_clicked)
a87.on_click(on_a87_clicked)
a88.on_click(on_a88_clicked)
a89.on_click(on_a89_clicked)
a90.on_click(on_a90_clicked)
In[9]: # Defining the gui widgets
selectAtom = widgets.Dropdown(
options= elements,
value=2,
description='Atom:',
)
atomsLabel = widgets.Checkbox(
value=False,
description='Atomic label',
disabled=False,
indent=False
)
emissionType= widgets.Dropdown(
options=['plane', 'cones'],
value='plane',
description='Emission:',
)
coloredEmissions = widgets.Checkbox(
value=False,
description='Color emissions',
disabled=False,
indent=False,
)
# Changes the equatorial(or cone) emission radius
emRSlider = widgets.FloatSlider(
value = 5.0,
min = 2.0,
max = 8.0,
step= 0.1,
description='EmRadius',
)
# Changes the distance between adjacent orthogonal protons
ePSlider = widgets.FloatSlider(
value = 5.0,
min = 2.0,
max = 7.0,
step= 0.1,
description='Equ1_Pole2',
)
# Changes the distance tween top and bottom alpha parallel protons
pPSlider = widgets.FloatSlider(
value = 4.0,
min = 3.0,
max = 6.0,
step= 0.1,
description='A width',
)
# Changes the pole to pole distance between 2 adjacent alphas
p2p3Slider = widgets.FloatSlider(
value = 3.0,
min = 2.0,
max = 5.0,
step= 0.1,
description='A1_A2 Sep',
)
# Changes the cone emission angle
coneAngleSlider = widgets.FloatSlider(
value = 30, # deg
min = 20,
max = 40,
step= 0.1,
description='Cone angle',
)
# The guiwidget control panel
guiwidgets = widgets.VBox([selectAtom,atomsLabel,emissionType, # moved
coloredEmissions,emRSlider,ePSlider,
pPSlider,p2p3Slider,coneAngleSlider])
# Observe any changes in widget values, button or gui
number = selectAtom.value
def handle_change(change):
global number
number = change.new
selectAtom.observe(handle_change, 'value')
emR = emRSlider.value
def handle_change(change):
global emR
emR = change.new
emRSlider.observe(handle_change, 'value');
ortho = ePSlider.value # proton1 equator to proton2 pole
def handle_change(change):
global ortho
ortho = change.new
ePSlider.observe(handle_change, 'value');
pPTP = pPSlider.value # proton pole to proton pole
def handle_change(change):
global pPTP
pPTP = change.new
pPSlider.observe(handle_change, 'value')
p2p3Dist = p2p3Slider.value # between 2 adjacent alphas
def handle_change(change):
global p2p3Dist
p2p3Dist = change.new
p2p3Slider.observe(handle_change, 'value')
cAngleDeg = coneAngleSlider.value # cone angle
def handle_change(change):
global cAngleDeg
cAngleDeg = change.new
coneAngleSlider.observe(handle_change, 'value')
In[10]: grid # Display the Periodic Table
In[11]: guiwidgets # Display the main control widgets.
In[12]: # After making an element change, re-run this cell
#and the rest.
An = elements[number-1]
atomicLabel = An[0]
atomicLabel
Last time I announced milestone progress. A lengthy, explicit 90 tuple list - 'elements', was replaced with a much shorter coded tuple list entirely derived from Cr6-Elements.csv. ‘elements’ was working properly, identifying desired atom changes with either dropdown or Periodic table selections. Only one problem, I had to use (' hydrogen', 1), instead of the intended contents of the ‘AtomicTuple’ column, with values like (' 1 hydrogen H ', 1).
1. That problem has been corrected, the program now includes the ‘AtomicTuple’ column values as intended. The code read each ‘AtomicTuple’ value as a string - not a tuple. After a google search I found one may convert a string to a tuple by a combined use of list, map and eval methods as shown below, also included in the attached code, In[7].
- Code:
atomicTups = df['AtomicTuple'].drop_duplicates().array
elements = list(map(eval, atomicTups))
2. In the code section’s In[8] the periodic table buttons are defined, using several other long written lists. ‘dlist’, a list of the atomic symbols ( H through Th) is now also derived from the .csv file with the pandas code:
- Code:
dlist = df['AtomicSymbol'].drop_duplicates().array
Next I decided to go after clist = [clrs[5],clrs[7],clrs[0], … a 90 item list which is used to color each periodic table entry according to the atom’s type: alkali metal, alkaline earth, … , noble gas. See In[8] for the 10 types and the ‘clrs’ list of 10 colors.
3. Added a new column to Cr6-Elements.csv, ‘AtomicType’ that will help me cleanup the lists In[8].
Since then I’ve reviewed python lists, tuples and dictionaries. I’ll see what happens next.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Happy to report a little more progress.
1. Creating ‘clist’ at the top of In[8]: # The periodic table's 90 buttons ...” has been changed to the following code.
- Code:
# In[8]: The periodic table's 90 buttons. Each button includes:
# Atomic type color, 'clist', worked in steps first-fifth. Plus an
# Atomic symbol, dlist; and button layout width and height.
# First, the ten Atomic types, tenATypes.
# tenATypes = list(df['Atomic ... works ok, the explicit list is simpler,
# as previous atom selection data values are over-written.
#tenATypes = []
#tenATypes = list(df['AtomicType'].drop_duplicates()) # 1710 rows down to 10 rows
tenATypes = ['nonmetal','noble_gas','alkali_metal','alkaline_earth','metalloid',
'halogen','basic_metal','transition_metal','lanthanide','actinide']
# Second - The 10 atomic type colors, tenColors. Lined up to agree with tenATypes
tenColors = ['skyblue','cyan','tomato','orange','springgreen','powderblue','palegreen',
'yellow','thistle','violet']
# Third, zip together the type/color dictionary
typeColorDict = {}
typeColorDict = dict(zip(tenATypes, tenColors))
#typeColorDict
# Fourth - creating an ('Atomicnumber','AtomicType')
# a 90 item tuple list.
df['allATypes'] = list(zip(df.AtomicNumber, df.AtomicType))
ATypes = df['allATypes'].drop_duplicates().array
ATypes
# Fifth - Build the list of 90 colors, clist
clist = []
for i in range(90):
clist.append(typeColorDict[ATypes[i][1]])
#clist
# The atomic symbols for the Periodic table.
dlist = df['AtomicSymbol'].drop_duplicates().array
# ...

2. Copied the SlotLayout cells from Atom Builder to mBuilder and changed the SL slot numbers and orientations to agree with the SL configuration in Cr6-Elements.csv. In AB, the slotlist, a set of 19 integers identifying the number of protons in each numbered slot, was created in the 19 loops. It should be a lot easier to simply read the slot totals from the .csv file. The slotlist for Neon is [2,2,2,2,2,0,0,0,0,0,0,0,0,0,0,0,0,0,0].
3. df["AtomicNumber"] = df["AtomicNumber"].astype(str)
I wasn’t able to cleanup the Periodic table lists as much as I’d hoped. Both the Periodic table buttons as well as the SL buttons use df["AtomicNumber"]. I decided to convert "AtomicNumber" from an integer to a string type data, I think that might help.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Just wanted to "dangle" this as a possible place to look at in terms of the photon. Graph Networkx with Mathis' photonic structures. Heavy duty algos here but there is something that might allow for "molecular-bonds" with via Miles' slots-stacked spins with Graph coding. I don't want to push this too hard and it may need it's own "thread" on the forum. It's hot stuff at Stanford, MIT, Harvard for shaping "relationships"...for us it is bonds with atoms on slots. The look at "weights" between edges and nodes. There might be relationships that can be leveraged-ML'd with the weights that can be assigned for the C.F. per Mathis. It might be good at saying these atoms can bond given the Periodic Table (and C.F. flows per Mathis)...like which bonds can be formed with these stand-alone atoms using the Graphs?:
https://web.stanford.edu/class/cs224w/slides/08-GNN-application.pdf
https://snap.stanford.edu/pgnn/#datasets
Notebook examples: https://github.com/snap-stanford/GraphGym
The paper that kicked this off in high gear a few years ago: https://arxiv.org/pdf/1906.04817v2.pdf
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
- Code:
In[12]: # After making an element change, re-run this cell
#and the rest.
An = elements[number-1]
atomicLabel = An[0]
atomicNumber = number
# The slotlist
rowstart = 19 * (atomicNumber - 1)
rowstop = rowstart + 19
slotlist = list(df.iloc[rowstart:rowstop, 14])
atomicLabel
In[13]: # Create the Slotlayout Diagram.
# The slotlist, a set of 19 integers identifying
# the number of protons in each numbered slot is
# found in the df['AlphaType'] column, (14). and
# is identified in the previous cell.
# Using 45 non-interactive buttons to create the
# atom's slotlayout (19 + 19 + 7 + 1) diagram: 19
# in the center SL configuration; 19 for the left
# (1,3,5,...,19) and right(2,4,6,...,18) columns;
# 7 for the slot/proton count color legend at the
# bottom; and 1 atomic symbol/mumber at the bottom
# right. All buttons except the bottom right incl:
# proton count color, layout width and layout height.
# The 7 colors used for color-coding each slot's
# total proton counts, 0-6.
sclrs = ['white','lightgray','dodgerblue','pink','salmon','greenyellow','skyblue']
sdlist = slotlist
# The colors for the 19 slots, sclist, according to
# the number of protons in each.
sclist = np.zeros(19, dtype='object')
for i in range(19):
sclist[i] = sclrs[slotlist[i]]
# Defining the slot buttons by adding the slot position, count and colors
s1 = Button(description='1 --- '+str(sdlist[0]), style=dict(button_color=sclist[0]))
s2 = Button(description='2 | '+str(sdlist[1]), style=dict(button_color=sclist[1]))
s3 = Button(description='3 | '+str(sdlist[2]), style=dict(button_color=sclist[2]))
s4 = Button(description='4 --- '+str(sdlist[3]), style=dict(button_color=sclist[3]))
s5 = Button(description='5 --- '+str(sdlist[4]), style=dict(button_color=sclist[4]))
s6 = Button(description='6 | '+str(sdlist[5]), style=dict(button_color=sclist[5]))
s7 = Button(description='7 | '+str(sdlist[6]), style=dict(button_color=sclist[6]))
s8 = Button(description='8 | '+str(sdlist[7]), style=dict(button_color=sclist[7]))
s9 = Button(description='9 | '+str(sdlist[8]), style=dict(button_color=sclist[8]))
s10 = Button(description='10 -- '+str(sdlist[9]), style=dict(button_color=sclist[9]))
s11 = Button(description='11 -- '+str(sdlist[10]), style=dict(button_color=sclist[10]))
s12 = Button(description='12 -- '+str(sdlist[11]), style=dict(button_color=sclist[11]))
s13 = Button(description='13 -- '+str(sdlist[12]), style=dict(button_color=sclist[12]))
s14 = Button(description='14 | '+str(sdlist[13]), style=dict(button_color=sclist[13]))
s15 = Button(description='15 | '+str(sdlist[14]), style=dict(button_color=sclist[14]))
s16 = Button(description='16 -- '+str(sdlist[15]), style=dict(button_color=sclist[15]))
s17 = Button(description='17 -- '+str(sdlist[16]), style=dict(button_color=sclist[16]))
s18 = Button(description='18 -- '+str(sdlist[17]), style=dict(button_color=sclist[17]))
s19 = Button(description='19 -- '+str(sdlist[18]), style=dict(button_color=sclist[18]))
# Slotlayout proton total color legend
s20 = Button(description=str('0'), style=dict(button_color=sclrs[0]))
s21 = Button(description=str('1'), style=dict(button_color=sclrs[1]))
s22 = Button(description=str('2'), style=dict(button_color=sclrs[2]))
s23 = Button(description=str('3'), style=dict(button_color=sclrs[3]))
s24 = Button(description=str('4'), style=dict(button_color=sclrs[4]))
s25 = Button(description=str('5'), style=dict(button_color=sclrs[5]))
s26 = Button(description=str('6'), style=dict(button_color=sclrs[6]))
# Left and right slot columns,
s27 = Button(description='1 --- '+str(sdlist[0]), style=dict(button_color=sclist[0]))
s28 = Button(description='2 | '+str(sdlist[1]), style=dict(button_color=sclist[1]))
s29 = Button(description='3 | '+str(sdlist[2]), style=dict(button_color=sclist[2]))
s30 = Button(description='4 --- '+str(sdlist[3]), style=dict(button_color=sclist[3]))
s31 = Button(description='5 --- '+str(sdlist[4]), style=dict(button_color=sclist[4]))
s32 = Button(description='6 | '+str(sdlist[5]), style=dict(button_color=sclist[5]))
s33 = Button(description='7 | '+str(sdlist[6]), style=dict(button_color=sclist[6]))
s34 = Button(description='8 | '+str(sdlist[7]), style=dict(button_color=sclist[7]))
s35 = Button(description='9 | '+str(sdlist[8]), style=dict(button_color=sclist[8]))
s36 = Button(description='10 -- '+str(sdlist[9]), style=dict(button_color=sclist[9]))
s37 = Button(description='11 -- '+str(sdlist[10]), style=dict(button_color=sclist[10]))
s38 = Button(description='12 -- '+str(sdlist[11]), style=dict(button_color=sclist[11]))
s39 = Button(description='13 -- '+str(sdlist[12]), style=dict(button_color=sclist[12]))
s40 = Button(description='14 | '+str(sdlist[13]), style=dict(button_color=sclist[13]))
s41 = Button(description='15 | '+str(sdlist[14]), style=dict(button_color=sclist[14]))
s42 = Button(description='16 -- '+str(sdlist[15]), style=dict(button_color=sclist[15]))
s43 = Button(description='17 -- '+str(sdlist[16]), style=dict(button_color=sclist[16]))
s44 = Button(description='18 -- '+str(sdlist[17]), style=dict(button_color=sclist[17]))
s45 = Button(description='19 -- '+str(sdlist[18]), style=dict(button_color=sclist[18]))
# The slotlayout atom
s46 = Button(description=dlist[number-1]+' '+str(number), style=dict(button_color=sclrs[0],font_weight='bold'))
# Creating the list of buttons: 19,7,19 and 1.
sbttns = [s1,s2,s3,s4,s5,s6,s7,s8,s9,s10,s11,s12,s13,s14,s15,s16,s17,s18,s19,
s20,s21,s22,s23,s24,s25,s26,s27,s28,s29,s30,s31,s32,s33,s34,s35,s36,
s37,s38,s39,s40,s41,s42,s43,s44,s45,s46]
# Add each button's border, layout width and height.
i=0
while i < 46 :
sbttns[i].layout = widgets.Layout(border = '1px solid black')
sbttns[i].layout.width = '100%'
sbttns[i].layout.height = '100%'
i += 1
# Layout the 46 (19 + 19 + 7 + 1) slotlayout buttons
sgrid = GridspecLayout(10, 9)
sgrid[1,4] = sbttns[13] # Cr6-Elements.csv SlotLayout.
sgrid[2,4] = sbttns[3]
sgrid[2,5] = sbttns[8]
sgrid[2,6] = sbttns[12]
sgrid[3,3] = sbttns[17]
sgrid[3,4] = sbttns[1]
sgrid[3,5] = sbttns[15]
sgrid[4,2] = sbttns[9]
sgrid[4,3] = sbttns[5]
sgrid[4,4] = sbttns[0]
sgrid[4,5] = sbttns[6]
sgrid[4,6] = sbttns[11]
sgrid[5,3] = sbttns[16]
sgrid[5,4] = sbttns[2]
sgrid[5,5] = sbttns[18]
sgrid[6,2] = sbttns[10]
sgrid[6,3] = sbttns[7]
sgrid[6,4] = sbttns[4]
sgrid[7,4] = sbttns[14]
sgrid[9,1] = sbttns[19] # Color/num protons legend
sgrid[9,2] = sbttns[20] # along the bottom row
sgrid[9,3] = sbttns[21]
sgrid[9,4] = sbttns[22]
sgrid[9,5] = sbttns[23]
sgrid[9,6] = sbttns[24]
sgrid[9,7] = sbttns[25]
sgrid[0,0] = sbttns[26] # L/R Vertical slot columns
sgrid[0,8] = sbttns[27]
sgrid[1,0] = sbttns[28]
sgrid[1,8] = sbttns[29]
sgrid[2,0] = sbttns[30]
sgrid[2,8] = sbttns[31]
sgrid[3,0] = sbttns[32]
sgrid[3,8] = sbttns[33]
sgrid[4,0] = sbttns[34]
sgrid[4,8] = sbttns[35]
sgrid[5,0] = sbttns[36]
sgrid[5,8] = sbttns[37]
sgrid[6,0] = sbttns[38]
sgrid[6,8] = sbttns[39]
sgrid[7,0] = sbttns[40]
sgrid[7,8] = sbttns[41]
sgrid[8,0] = sbttns[42]
sgrid[8,8] = sbttns[43]
sgrid[9,0] = sbttns[44]
sgrid[9,8] = sbttns[45] # Atomic symbpl and number
In[14]: The Slot Layout Diagram.
All atoms can be described by the number of protons (0-6)
in each of the atom's (1-19) occupied slot positions.
The main vertical up/down channel slots (top to bottom) are
(14,4,2,1,3,5,15). There is also the front/back channel
(11,17,1,16,13), and left/right channel (10,6,1,7,12); or
refer to the directions from center slot 1: +/-x, +/-y, and
main vertical column +/-z. The front/back/ left/right arms
spin as a single group, the Carousel, around slot 1. Hydroden
has a single proton in its single occupied slot 1. Helium's
single slot 1 contains two protons. Hook slot positions:
16,17,18, and 19, are joined to the main up/down column at
slots, 2 and 3.
All but most of the bottom row buttons contain number pairs
separated by either | or --, the vertical or horizontal
characters indicate the direction of that slot's proton
equatorial emission planes viewed edgewise, orthogonal to
the proton's spin axis. The left side number indicates the
1-19 slot number. The right side number indicates the number
of protons in that atom's slot.
The Slot Layout is shown in the atomic configuration in the
center of the diagram. It is a 3D, six pointed star projected
onto the diagram's flat surface. The slot button information
is repeated in the left and right side vertical columns for
convienence. The seven center buttons in the bottom row
contains the total protons per slot color legend. The selected
atom’s symbol and atomic number are in the bottom right corner.
In[15]: sgrid # Display the atom's slotlayout diagram
In[16]:# Build atomic models.
grpAn = Group() # The rotation widget spins grpAn, slots 1,4 and 5.
grpBn = Group() # grpBn are the orthogonal top, bottom and hook slots.
proStack1 = Group() # Pulled out of the i-loop after
proStack2 = Group() # proStack not defined errors.
proStack3 = Group()
proStack4 = Group()
proStack5 = Group()
proStack6 = Group()
proStack7 = Group()
proStack8 = Group()
proStack9 = Group()
proStack10 = Group()
proStack11 = Group()
proStack12 = Group()
proStack13 = Group()
proStack14 = Group()
proStack15 = Group()
proStack16 = Group()
proStack17 = Group()
proStack18 = Group()
proStack19 = Group()
# Dimensions # pRadius is now in In[1]
nRadius = pRadius*1.125 # Neutron radius
eRadius = 2*pRadius/10 # Electron radius
# Widget Controlled variables 1. ortho, 2.pPTP, 3. p2p3Dist, 4. cAngleDeg
orthoP = ortho*pRadius
emRadius = 2*emR*pRadius # 2*orthoP - pRadius/2
coneAngle = cAngleDeg*2*piG/360
coneHeight = emRadius*tan(coneAngle)
pP1pP2 = 2*pPTP*pRadius # Widget control.
p2p3 = 2*p2p3Dist*pRadius # Widget control
n1n2 = (pP1pP2- 2*nRadius)/tan(coneAngle) # neutron-neutron orbital separation (y) inside the alpha
# e1e2 = -n1n2/10
p1p2 = pRadius*2.5 #0.75*pP1pP2 # pRadius*2.5
p1n1 = pP1pP2/2 #pP1pP2/2 #pP1pP2/2 # Alpha, up-down (z) proton-orbital neutron separation
p1n2 = pP1pP2/2 #pRadius + nRadius + 1.5*pRadius # In line proton and neutron
p1e1 = pRadius + 0.05*pP1pP2 # Alpha, top-bottom (z) proton-electron separation
maxZNeg = 0.0 # maxZNeg and maxZPos are Used to center(z) the atom on the screen
maxZPos = 0.0
# Proton set Geometries and Materials
protonGeometry = SphereBufferGeometry(pRadius, 24, 16)
neutronGeometry = SphereBufferGeometry(nRadius, 24, 16)
electronGeometry = SphereBufferGeometry(eRadius, 8, 16)
emissionGeometryP = CircleGeometry(emRadius, 24)
emissionGeometryX = ConeGeometry( emRadius, coneHeight, 16, openEnded = True )
protonMaterial = MeshStandardMaterial(color='blue')
protonMaterialB = MeshStandardMaterial(color='green')
neutronMaterial = MeshStandardMaterial(color='red')
neutronMaterialB = MeshStandardMaterial(color='green') # Color.js Find a nice color spectrum
electronMaterial = MeshStandardMaterial(color='cyan') #'orangered','salmon','green','cyan','teal','orange'
emissionmaterial7= MeshStandardMaterial(color='gray',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial1 = MeshStandardMaterial(color='magenta',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial2 = MeshStandardMaterial(color='blue',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial3 = MeshStandardMaterial(color='pink',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial4 = MeshStandardMaterial(color='salmon',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial5 = MeshStandardMaterial(color='lime',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial6 = MeshStandardMaterial(color='skyblue',side='DoubleSide',transparent = True,opacity = 0.20)
def buildProtonStack(a,j): # a is the total # of protons. j indicates the calling register
""" The odd 'a' returns just the single, first, third, or fifth proton. The even 'a'
returns the first, second or third alpha set, each 2 protons and 2 neutrons. """
num = 0
if coloredEmissions.value == True:
if a == 1 : emissionmaterial = emissionmaterial1
if a == 2 : emissionmaterial = emissionmaterial2
if a == 3 : emissionmaterial = emissionmaterial3
if a == 4 : emissionmaterial = emissionmaterial4
if a == 5 : emissionmaterial = emissionmaterial5
if a == 6 : emissionmaterial = emissionmaterial6
else:
emissionmaterial = emissionmaterial7
# Proton z positions: # The possible formula solution, rather than the list, may be in these 2 lines
# protonMi.position = [0,0,a*pP1pP2/2+(a-1)*p2p3/2-(p2p3/2+pP1pP2/2)*num] # multiAlpha solution - Not
# protonMi.position = [0,0,(a//2)*pP1pP2/2+(a//2)*p2p3/2-(p2p3/2+pP1pP2/2)*num] # pSet
if a == 1 : pPositions = [0]
if a == 2 : pPositions = [pP1pP2/2,-pP1pP2/2]
if a == 3 : pPositions = [pP1pP2/2+p2p3/2,-pP1pP2/2+p2p3/2,-pP1pP2/2-p2p3/2]
if a == 4 : pPositions = [pP1pP2+p2p3/2,p2p3/2,-p2p3/2,-pP1pP2-p2p3/2]
if a == 5 : pPositions = [pP1pP2+p2p3,p2p3,0,-pP1pP2,-pP1pP2-p2p3]
if a == 6 : pPositions = [3*pP1pP2/2+p2p3,pP1pP2/2+p2p3,pP1pP2/2,-pP1pP2/2,-pP1pP2/2-p2p3,-3*pP1pP2/2-p2p3]
while num < a:
protonMi = Group()
protonMi.add(Mesh(protonGeometry, protonMaterial))
if emissionType.value == 'plane':
protonMi.add(Mesh(emissionGeometry, emissionmaterial))
if emissionType.value == 'cones':
emission1 = Object3D()
emission1.add(Mesh(emissionGeometry, emissionmaterial))
emission1.position = [0,0,coneHeight/2]
emission1.rotateX(-piG/2)
protonMi.add(emission1)
emission2 = Object3D()
emission2.add(Mesh(emissionGeometry, emissionmaterial))
emission2.position = [0,0,-coneHeight/2]
emission2.rotateX(piG/2)
protonMi.add(emission2)
protonMi.add(Mesh(emissionGeometryP, emissionmaterial))
electronMi = Object3D()
electronMi.add(Mesh(electronGeometry, electronMaterial))
electronMi.position = [0,-n1n2/10,-p1e1]
protonMi.add(electronMi)
protonMi.rotateZ(random()*2*piG)
if num%2 != 0:
neutronPair = Group()
neutronMi = Object3D()
neutronMi.add(Mesh(neutronGeometry, neutronMaterial))
neutronMi.position = [0,-n1n2/2,-p1n1]
neutronNi = Object3D()
neutronNi.add(Mesh(neutronGeometry, neutronMaterial))
neutronNi.position = [0,n1n2/2,-p1n1]
neutronPair.add(neutronMi)
neutronPair.add(neutronNi)
neutronPair.rotateZ(random()*2*piG)
protonMi.add(neutronPair)
protonMi.rotateX(piG)
protonMi.rotateZ(random()*2*piG)
protonMi.position = [0,0,pPositions[num]]
if j == 0 : proStack1.add(protonMi)
if j == 1 : proStack2.add(protonMi)
if j == 2 : proStack3.add(protonMi)
if j == 3 : proStack4.add(protonMi)
if j == 4 : proStack5.add(protonMi)
if j == 5 : proStack6.add(protonMi)
if j == 6 : proStack7.add(protonMi)
if j == 7 : proStack8.add(protonMi)
if j == 8 : proStack9.add(protonMi)
if j == 9 : proStack10.add(protonMi)
if j == 10 : proStack11.add(protonMi)
if j == 11 : proStack12.add(protonMi)
if j == 12 : proStack13.add(protonMi)
if j == 13 : proStack14.add(protonMi)
if j == 14 : proStack15.add(protonMi)
if j == 15 : proStack16.add(protonMi)
if j == 16 : proStack17.add(protonMi)
if j == 17 : proStack18.add(protonMi)
if j == 18 : proStack19.add(protonMi)
num += 1
return
"""
# Assemble the atomic models.
for i in range( atomicNumber ):
if emissionType.value == 'plane':
emissionGeometry = emissionGeometryP
if emissionType.value == 'cones':
emissionGeometry = emissionGeometryX
if coloredEmissions.value == True:
emissionmaterial = emissionmaterial1
else:
emissionmaterial = emissionmaterial7
neutronGi = Object3D()
neutronGi.add(Mesh(neutronGeometry, neutronMaterial))
neutronKi = Object3D()
neutronKi.add(Mesh(neutronGeometry, neutronMaterial))
neutronLi = Object3D()
neutronLi.add(Mesh(neutronGeometry, neutronMaterial))
if i == 0: # Slot 1
if 1 == atomicNumber: # Hydrogen. Slot 1
buildProtonStack(1,i)
neutronGi.position = [0,0,-p1n2]
proStack1.position = [0, 0, 0]
proStack1.add(neutronGi)
grpAn.add(proStack1)
slotlist[i] = 1
if 2 <= atomicNumber < 6: # Helium
buildProtonStack(2,i)
grpAn.add(proStack1)
slotlist[i] = 2
if 6 == atomicNumber :
buildProtonStack(4,i)
grpAn.add(proStack1)
slotlist[i] = 4
maxZNeg = -3*pP1pP2/2-2*orthoP
maxZPos = 3*pP1pP2/2+2*orthoP
if 7 <= atomicNumber < 10 :
buildProtonStack(6,i)
grpAn.add(proStack1)
slotlist[i] = 6
maxZNeg = -3*pP1pP2/2-p2p3
maxZPos = p2p3+3*pP1pP2/2+2*orthoP
if 10 <= atomicNumber < 36 :
buildProtonStack(2,i)
grpAn.add(proStack1)
slotlist[i] = 2
if 36 <= atomicNumber < 54 :
buildProtonStack(4,i)
grpAn.add(proStack1)
slotlist[i] = 4
if 54 <= atomicNumber < 57:
buildProtonStack(6,i)
grpAn.add(proStack1)
slotlist[i] = 6
if 57 <= atomicNumber < 71:
buildProtonStack(5,i)
grpAn.add(proStack1)
slotlist[i] = 5
if 71 <= atomicNumber :
buildProtonStack(6,i)
grpAn.add(proStack1)
slotlist[i] = 6
"""
print(atomicNumber)
print(slotlist)
In[17]:# The widget tab enclosure.
# Including a non-interactive periodic table showing the atom selected.
b1 = Button(description=dlist[0], style=dict(button_color=clist[0]))
b2 = Button(description=dlist[1], style=dict(button_color=clist[1]))
b3 = Button(description=dlist[2], style=dict(button_color=clist[2]))
b4 = Button(description=dlist[3], style=dict(button_color=clist[3]))
b5 = Button(description=dlist[4], style=dict(button_color=clist[4]))
b6 = Button(description=dlist[5], style=dict(button_color=clist[5]))
b7 = Button(description=dlist[6], style=dict(button_color=clist[6]))
b8 = Button(description=dlist[7], style=dict(button_color=clist[7]))
b9 = Button(description=dlist[8], style=dict(button_color=clist[8]))
b10 = Button(description=dlist[9], style=dict(button_color=clist[9]))
b11 = Button(description=dlist[10], style=dict(button_color=clist[10]))
b12 = Button(description=dlist[11], style=dict(button_color=clist[11]))
b13 = Button(description=dlist[12], style=dict(button_color=clist[12]))
b14 = Button(description=dlist[13], style=dict(button_color=clist[13]))
b15 = Button(description=dlist[14], style=dict(button_color=clist[15]))
b16 = Button(description=dlist[15], style=dict(button_color=clist[15]))
b17 = Button(description=dlist[16], style=dict(button_color=clist[16]))
b18 = Button(description=dlist[17], style=dict(button_color=clist[17]))
b19 = Button(description=dlist[18], style=dict(button_color=clist[18]))
b20 = Button(description=dlist[19], style=dict(button_color=clist[19]))
b21 = Button(description=dlist[20], style=dict(button_color=clist[20]))
b22 = Button(description=dlist[21], style=dict(button_color=clist[22]))
b23 = Button(description=dlist[22], style=dict(button_color=clist[22]))
b24 = Button(description=dlist[23], style=dict(button_color=clist[23]))
b25 = Button(description=dlist[24], style=dict(button_color=clist[24]))
b26 = Button(description=dlist[25], style=dict(button_color=clist[25]))
b27 = Button(description=dlist[26], style=dict(button_color=clist[26]))
b28 = Button(description=dlist[27], style=dict(button_color=clist[27]))
b29 = Button(description=dlist[28], style=dict(button_color=clist[28]))
b30 = Button(description=dlist[29], style=dict(button_color=clist[29]))
b31 = Button(description=dlist[30], style=dict(button_color=clist[30]))
b32 = Button(description=dlist[31], style=dict(button_color=clist[31]))
b33 = Button(description=dlist[32], style=dict(button_color=clist[32]))
b34 = Button(description=dlist[33], style=dict(button_color=clist[33]))
b35 = Button(description=dlist[34], style=dict(button_color=clist[34]))
b36 = Button(description=dlist[35], style=dict(button_color=clist[35]))
b37 = Button(description=dlist[36], style=dict(button_color=clist[36]))
b38 = Button(description=dlist[37], style=dict(button_color=clist[37]))
b39 = Button(description=dlist[38], style=dict(button_color=clist[38]))
b40 = Button(description=dlist[39], style=dict(button_color=clist[39]))
b41 = Button(description=dlist[40], style=dict(button_color=clist[40]))
b42 = Button(description=dlist[41], style=dict(button_color=clist[41]))
b43 = Button(description=dlist[42], style=dict(button_color=clist[42]))
b44 = Button(description=dlist[43], style=dict(button_color=clist[43]))
b45 = Button(description=dlist[44], style=dict(button_color=clist[44]))
b46 = Button(description=dlist[45], style=dict(button_color=clist[45]))
b47 = Button(description=dlist[46], style=dict(button_color=clist[46]))
b48 = Button(description=dlist[47], style=dict(button_color=clist[47]))
b49 = Button(description=dlist[48], style=dict(button_color=clist[48]))
b50 = Button(description=dlist[49], style=dict(button_color=clist[49]))
b51 = Button(description=dlist[50], style=dict(button_color=clist[50]))
b52 = Button(description=dlist[51], style=dict(button_color=clist[51]))
b53 = Button(description=dlist[52], style=dict(button_color=clist[52]))
b54 = Button(description=dlist[53], style=dict(button_color=clist[53]))
b55 = Button(description=dlist[54], style=dict(button_color=clist[54]))
b56 = Button(description=dlist[55], style=dict(button_color=clist[55]))
b57 = Button(description=dlist[56], style=dict(button_color=clist[56]))
b58 = Button(description=dlist[57], style=dict(button_color=clist[57]))
b59 = Button(description=dlist[58], style=dict(button_color=clist[58]))
b60 = Button(description=dlist[59], style=dict(button_color=clist[59]))
b61 = Button(description=dlist[60], style=dict(button_color=clist[60]))
b62 = Button(description=dlist[61], style=dict(button_color=clist[61]))
b63 = Button(description=dlist[62], style=dict(button_color=clist[62]))
b64 = Button(description=dlist[63], style=dict(button_color=clist[63]))
b65 = Button(description=dlist[64], style=dict(button_color=clist[64]))
b66 = Button(description=dlist[65], style=dict(button_color=clist[65]))
b67 = Button(description=dlist[66], style=dict(button_color=clist[66]))
b68 = Button(description=dlist[67], style=dict(button_color=clist[67]))
b69 = Button(description=dlist[68], style=dict(button_color=clist[68]))
b70 = Button(description=dlist[69], style=dict(button_color=clist[69]))
b71 = Button(description=dlist[70], style=dict(button_color=clist[70]))
b72 = Button(description=dlist[71], style=dict(button_color=clist[71]))
b73 = Button(description=dlist[72], style=dict(button_color=clist[72]))
b74 = Button(description=dlist[73], style=dict(button_color=clist[73]))
b75 = Button(description=dlist[74], style=dict(button_color=clist[74]))
b76 = Button(description=dlist[75], style=dict(button_color=clist[75]))
b77 = Button(description=dlist[76], style=dict(button_color=clist[76]))
b78 = Button(description=dlist[77], style=dict(button_color=clist[77]))
b79 = Button(description=dlist[78], style=dict(button_color=clist[78]))
b80 = Button(description=dlist[79], style=dict(button_color=clist[79]))
b81 = Button(description=dlist[80], style=dict(button_color=clist[80]))
b82 = Button(description=dlist[81], style=dict(button_color=clist[81]))
b83 = Button(description=dlist[82], style=dict(button_color=clist[82]))
b84 = Button(description=dlist[83], style=dict(button_color=clist[83]))
b85 = Button(description=dlist[84], style=dict(button_color=clist[84]))
b86 = Button(description=dlist[85], style=dict(button_color=clist[85]))
b87 = Button(description=dlist[86], style=dict(button_color=clist[86]))
b88 = Button(description=dlist[87], style=dict(button_color=clist[87]))
b89 = Button(description=dlist[88], style=dict(button_color=clist[88]))
b90 = Button(description=dlist[89], style=dict(button_color=clist[89]))
# Creating the list of buttons
bbttns = [b1,b2,b3,b4,b5,b6,b7,b8,b9,b10,b11,b12,b13,b14,b15,b16,b17,b18,b19,
b20,b21,b22,b23,b24,b25,b26,b27,b28,b29,b30,b31,b32,b33,b34,b35,b36,b37,b38,b39,
b40,b41,b42,b43,b44,b45,b46,b47,b48,b49,b50,b51,b52,b53,b54,b55,b56,b57,b58,b59,
b60,b61,b62,b63,b64,b65,b66,b67,b68,b69,b70,b71,b72,b73,b74,b75,b76,b77,b78,b79,
b80,b81,b82,b83,b84,b85,b86,b87,b88,b89,b90]
bbttns[atomicNumber-1] = Button(description=dlist[atomicNumber-1], style=dict(button_color='white',font_weight='bold'))
# Use the button list in a loop to add each button's border, layout width and height.
i=0
while i < 90 :
if i == atomicNumber-1 :
bbttns[atomicNumber-1].layout = widgets.Layout(border = '2px solid red')
else :
bbttns[i].layout = widgets.Layout(border = '1px solid black')
bbttns[i].layout.width = '100%'
bbttns[i].layout.height = '100%' #'40px'
i += 1
# Layout the 90 buttons in periodic table form
gridb = GridspecLayout(10, 18)
gridb[0,0] = bbttns[0]
gridb[0,17] = bbttns[1]
gridb[1,0] = bbttns[2]
gridb[1,1] = bbttns[3]
p=12
while p < 18:
gridb[1,p] = bbttns[p-8]
p += 1
gridb[2,0] = bbttns[10]
gridb[2,1] = bbttns[11]
n=12
while n < 18:
gridb[2,n] = bbttns[n]
n += 1
m=0
while m < 18:
gridb[3,m] = bbttns[18+m]
m += 1
k=0
while k < 18:
gridb[4,k] = bbttns[36+k]
k += 1
gridb[5,0] = bbttns[54]
gridb[5,1] = bbttns[55]
# gridb[5,2] = the Lanthanide series hole
i=3
while i < 18:
gridb[5,i] = bbttns[68+i]
i += 1
gridb[6,0] = bbttns[86]
gridb[6,1] = bbttns[87]
# gridb[6,2] = the Actinide series hole
j=2
while j < 17:
# Lanthanide series
gridb[7,j] = bbttns[54+j]
j += 1
#Actinide series
gridb[8,2] = bbttns[88]
gridb[8,3] = bbttns[89]
# The tab enclosure
#tab1 = VBox(children=[renderer]) # 3D Atom
tab1 = VBox(children=[sgrid]) # Slotlayout diagram
#tab3 = VBox(children=[rgrid]) # Rotation controls
tab2 = VBox(children=[gridb]) # Periodic table
#tab = widgets.Tab(children=[tab1, tab2, tab3, tab4])
tab = widgets.Tab(children=[tab1, tab2])
#tab.set_title(0, atomicLabel)
tab.set_title(0, dlist[number-1]+' Slotlayout Diagram')
#tab.set_title(2, 'Slot Rotations')
tab.set_title(1, 'Periodic Table')
VBox(children=[tab]) # Display a tab enclosure with all outputs
In[17]:Charge Field physics and the Unified Field theory,
(and much more) have been well developed and well described by
Miles Mathis. See,
THE GREATEST STANDING ERRORS IN PHYSICS AND MATHEMATICS
http://milesmathis.com/index.html Miles Mathis science site
Homepage.
SECTION 9: THE NUCLEUS contains descriptions and diagrams of
charge channeling and charge recycling by the elements. A
paper most relevant to Atom Builder is
How to Build the Elements. Explaining the periodic table, with
nuclear diagrams".
http://milesmathis.com/nuclear.pdf How to Build the Elements
For Charge Field discussion visit the forum at "Miles Mathis'
Charge Field - Portal" https://milesmathis.forumotion.com.
This particular project is described in the Miles Periodic Table
with Standard Periodic Table reference thread,
https://milesmathis.forumotion.com/t634p75-miles-periodic-table-with-standard-periodic-table-reference#6702.
This project was Cr6's idea. He's also responsible for following
Miles Mathis' atomic model and creating the Slotlayout diagram
this project greatly relies.
There's at least one 10 year old charge field "Atom Viewer" out
there, Nevyn's object oriented, way sophisticated javaScript code.
I'll try to figure it out.
Please pardon my understandings, personal interpretations and
mistakes. Feel free to make this project better.
The code – From the atom selection/verification cell till the end.
I’m otherwise occupied tomorrow thru Monday, so here’s an early Project update.
1. The Periodic table (Pt) and SlotLayout (SL) diagram are reconfigured and properly read a working copy of Cr6-Elements.csv’s data file.
2. Added the tab enclosure which properly displays either a non-interactive Pt, or a second SL.
3. Added Atom Builder's 3D modeling cell's: Groups(), geometries, materials and functions necessary in order to begin making atoms.
I’ve replaced some lists with much shorter code; still, long lists remain, such as those associated with the SL and Pt’s 46 and 90 buttons. I’ll continue trying to figure out or find shorter alternatives.
I’m spending time wondering – not very focused – on how best to change the program flow. I suppose I should convert building both the SL and Pt into functions. That would make recreating them after selecting a new atom easier than repeating all the notebook cells beginning at the atom selection/verification cell as is currently done in AB.
Cr6, I’ve briefly looked at the Position-aware Graph Neural Network stuff. It almost reads like another language. For starters, I’d say, the number of points on the atom that can bond are limited, of course any node representing an atom should have positional awareness of those bonding points and directions.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
No atom building yet, but I’ve made plenty of preparatory changes:
1. Changed the values in Cr6-Elements.csv, ['OrbitalGroup'] column from objects: [Zero],[He],[Ne],[Ar],[Kr],[Xe],[Rn]; to integers: [0],[2],[10],[18],[36],[54],[86] in order to clear the Key Error:
In[]: df['OrbitalGroup'].describe()
Out []: KeyError: 'OrbitalGroup'.
2. Copied Cr6-Elements.csv [‘AlphaType’] column contents - indicating how many protons are in that slot, into the ‘protons’ column and moved ‘GroupNumber’ ‘Period’ and ‘SlotNumber’ columns to the left of ‘Protons’. Making it easier to reference the SlotLayout section data. The ‘AlphaType’ will be used to indicate how many alpha pairs are in that slot.
3. Added the last set of controls to mBuilder, the slot rotations. Those controls are included in a tab and they are also displayed just below the tab. View the atom tab and rotation controls to verify rotation changes as they are made, just as we first discussed what seems like a long time ago. A possible alternative, two tabs, one above the other, doesn’t work when both show the renderings, they bomb/disappear.
3. Combined two cells, df_raw and df, into one cell.
- Code:
In [12]: # Atom data. After making an element change,
# obtain sets of that atom's data.
# Selected Atom
An = elements[number-1]
atomicLabel = An[0]
atomicNumber = number
elementlist = df['Element'].drop_duplicates().array
atom = elementlist[number-1]
atomsymbol = dlist[number-1]
# print(atom, atomsymbol, atomicNumber)
# Selected Atom data; slotSet_df is a dataFrame
# containing the atom's SlotLayout details.
# slotset_df['Protons'] column and slotlist
# contain the number of protons in each slot.
rowstart = 19 * (atomicNumber - 1)
rowstop = rowstart + 19
slotset_df = df.iloc[rowstart:rowstop, 8:19].set_index('SlotNumber')
#slotset_df
#slotset_df.info()
slotlist = list(slotset_df.loc[:, 'Protons'])
# print(slotlist)
4. Added to the select/verification cell shown above, now starting with “# Atom Data” with new constants/variables 'atom', “atomsymbol”, and 'slotset_df' - an atomic dataframe with just 19 rows and SL data.
5. Moved build slotlayout (sgrid) cell to after the verification “Atom data” cell; sgrid and the rendered atom are now displayed in the tab enclosure.
I made several attempts to turn building sgrid, from the build slotlayout cell into a function without success.
The easiest way to run the program now is by highlighting the jupyter notebook atom verification/selection cell then select from the cell commands, “Run All Above”. Make any Atom or control panel changes, then from the cell commands select, “Run All Below”.
UserWarning. Shown above in pink, just after the renderer is defined. You may need to reduce the image size to see "UserWarning" cut off on the right side edge. Every time the program is first run, a UserWarning is displayed. The UserWarning is not displayed for additional runs, such as after selecting a new atom. I have no idea what the offending item is. The image shows where I’ve commented out the section responsible for adding the atom label to the 3D rendering, but that has no effect, the UserWarning is still displayed. After a search I’m afraid the warning is associated with the pythreejs renderer.
https://ipyvolume.readthedocs.io/en/latest/pythreejs.html
Has the same UserWarning, it’s shown below the 3D rendered rotating plot output below controls.
There’s no mention or discussion of the User Warning. Nor in a pdf copy of the same information at https://buildmedia.readthedocs.org/media/pdf/ipyvolume/latest/ipyvolume.pdf
See section 11.1 on doc page 64 (or pdf page 68), showing the same UserWarning, without the rendered image.
Otherwise, there's little left to do except to begin attempting to make dataframe atoms.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
https://stackoverflow.com/questions/38821132/bokeh-valueerror-out-of-range-float-values-are-not-json-compliant
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Cr6, This problem is present in both Atom Builder and mBuilder.
Thanks for the link, I read it the first time over a week ago. Even though I don’t have the 'bokeh' package I did a review of all the .csv and dataFrame df values for NaN’s. It all looked clean to me and the UserWarning was gone. I noticed the warning again, and thought it was an intermittent problem. I’d not yet realized the warning is only displayed on the program’s initial run.
This morning I searched on:
Jupyter User Warning: Message serialization failed with: Out of range float values are not JSON compliant
and found the github issue:
https://github.com/jupyter-widgets/pythreejs/issues/366
JSON serialization of OrbitControls fails with jupyter_client 7.0.3 #366
boehmc opened this issue on Sep 22, 2021 · 7 comments · Fixed by jupyter/jupyter_client#708
Updated my anaconda navigator from 1.1 to 1.2, and jupyter_client from 7.1.1 to 7.1.2.,
noting 708 doesn’t look a lot like 7.1.1 or 7.1.2 to me. Well that didn’t fix the problem.
Reading the github issue comments more closely it appears the problem lies in the fact that the pythreejs orbital camera’s built in values include a couple of JSON non-compliant infinite values, "float('inf')".
martinRenou commented on Sep 30, 2021
This will be fixed by jupyter/jupyter_client#708 (at least, we will only see a Warning but the widget will work again)
Though we will need to update pythreejs at some point, having a custom serializer to support Infinity/NaN, as it's not valid JSON, similar to what ipydatagrid does: https://github.com/bloomberg/ipydatagrid/blob/main/ipydatagrid/datagrid.py#L172-L178
So, that stink will continue until it is corrected by pythreejs. The UserWarning is just a warning, the widget operates properly. My current pythreejs version is 2.3.0 and my updated anaconda informs me there is no pythreejs updates currently available. I’ll add a note to both AB and mBuilder.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
js/scripts/three-class-config.js
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Attempting the link – js/scripts/three-class-config.js results in:
This site can’t be reached, js’s DNS address could not be found.
Diagnosing the problem.
I haven’t seen any discussion. The next3 links go to
“A Python / ThreeJS bridge for Jupyter Widgets”, at github.
https://github.com/jupyter-widgets/pythreejs
last Updated on Dec 6, 2021.
https://github.com/jupyter-widgets/pythreejs/blob/master/setup.py
Latest commit on Feb 22, 2021
https://github.com/jupyter-widgets/pythreejs/issues .
Switch to closed issues, note the last update on 30 Oct 21.
Yep, no activity since Nov/early Dec, is a bad sign. On the other hand I suppose gaps lasting several months probably aren’t unusual in the development business. I’ll keep an eye on it and mention it if I see any signs of life.
Organizing my thoughts, starting at near panic at an unsupported jupyter/pythreejs
bridge, of course I hope that ends up not being the case.
Ok, building atoms. I realized one of my main assumptions was wrong, I cannot simply populate each atomic slot’s x,y,z positions, because the atomic dimensions depend on user-controlled variables.
The basic atomic unit of measure the program uses is a non-user controllable proton radius, pRadius = 0.05.
1. emRadius, default(5) # Specifying the proton’s emission plane radius – doesn’t effect any atomic position values.
2. ortho, default(5). The distance between two orthogonal protons, a proton’s emission plane is aligned to an adjacent proton’s pole, the male-female type proton bond.
3. pPTP, default(4). # The distance between the centers of an Alpha’s two protons.
4. p2p3Dist, default(3). # The distance between the centers of the protons between 2 adjacent alphas. Also the distance between stacked (2-6) adjacent, parallel protons.
5. cAngleDeg, default(30deg). # define the latitude angle of the north/south cone emission maxima. Neutrons are placed so as to avoid the main +/- proton emission directions selected.
Those controls are not something I would choose to lose (funny rhyme/spelling that); however, maintaining that control capability makes things quite a bit more complicated than just reading a single dataFrame set, the program will often need to recalculate the slot positions before displaying an atom.
Still, its probably a good idea to load each of the 90 elements’ slot positional data with their initial unchanged control value distances/positions as the initial default value dataFrame position set. Perfectly good until you make a control distance value change, when the program will need to create a new slot position set – either each atom’s individual dataFrame or all 90 slot sets for each and every control distance value change. Creating new atomic position values is a necessary routine function.
Instead of transferring each positional float value by hand, as I’d originally imagined, I'll try transferring positional data to the dataFrame using mBuilder and the atom building section from Atom Builder. When the building cell sizes and positions each atomic particle or stack, the program should also save the position values in the 1710 row dataFrame df, within each atom’s up-to 19 row dataFrame, slotset_df.
So I set up the transition atom building cell, intended to write the default positional values into the Cr6-Elements.csv dataFrame. The atom building cell operates exactly like AB except all the other cells are mBuilder, using the 1710 row dataFrame df, and
each individual atomic dataframe, slotset_df. I imagine that after the initial dataframe is populated, it would be nice if the atom building cell transformed into the function which will create any new positional data sets as needed. Then a following section will
display either the default or newly calculated atomic positional data set.
Before I could go any further I needed to convert AB’s SlotLayout configuration into the mBuilder (csv file) SL configuration. For a fun problem, and an idea of the nuts and bolts I’m working with, having changed it in the first place, here are the steps I took, one pair switch at a time...
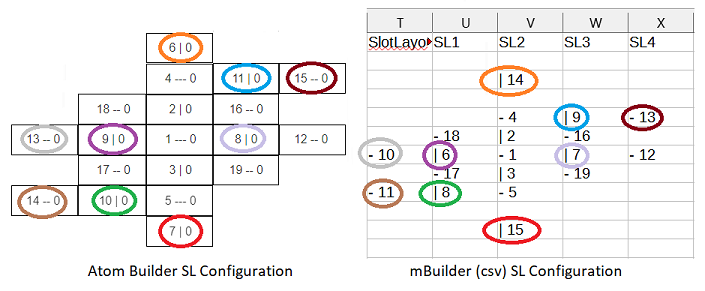
Going from the AB slotLayout configuration on the left to the mBuilder SL configuration on the right.
1. Put slot 14 where it belongs, switch slot 6 and slot 14 values. Slot 14 values are then in their correct positions.
2. Slot 6’s values now belong in slot 11’s position, switch slots 6 and 11 values. Slot 11 values are then in their correct positions.
3. Slots 6 and 9 values now belong in each other's positions, switch slots 6 and 9 values. Slots 6 and 9 values are then correct.
4. Give slot 15 slot 7’s values; switch slots 7 and 15. Slot 15 is then in its correct location.
5. Slot 7 values belong in slot 13’s positions, switch slots 7 and 13 slots. Slot 13 is then in its correct location.
6. Slot 7’s positions belong in slot 10, switch 7 and 10 slots. Slot 10 is then in its correct slot.
7. Slots 7 and 8 values belong in each other's positions. Switch Slots 7 and 8 values. All slots are then in their proper positions.
The slot 6 values changed and moved 3 times, and the slot 7 values changed/moved 4 times in order to reconfigure the SL. Of course each ‘pair switch’ is easier said than done. I usually involve a third unique ‘dummy’ placeholder. Ex. Switch f and n. Step 1. Switch ‘proStackf’ labels with ‘proStackQ’ labels. Step 2. Switch ‘proStackn’ labels to ‘proStackf’ labels. Step 3. Switch ‘proStackQ’ labels to ‘proStackn’ labels. Hard to believe I did all that without any major screw-ups, I’ll need to do a few verifications.
Correcting Atom Builder, 76, Osmium. Re-positioned the alpha in slot 18 (18 is not one of the reconfigured slots).
Uh oh, it appears all those PX, PY, PZ and NX, NY, NZ, 0.0 filled float values are immutable.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
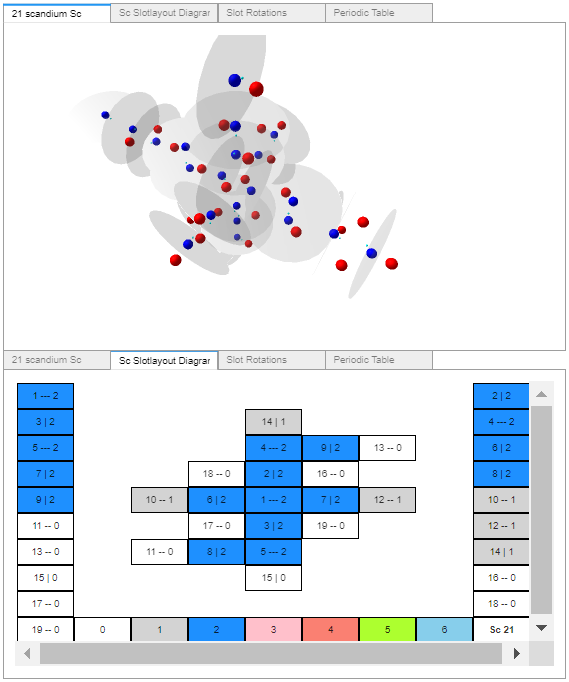
Significant progress. All Atom Builder’s 90 elements’ default proton and neutron coordinates have been transferred to Cr6-Elements.
Turns out the dataFrames’ x,y,z positional float values can be changed easily enough. Currently, when the atom is selected, all tabs operate normally. In addition, the updated slotset_df dataframe containing the proton and neutron coordinates for that atom is also displayed, as well as output to a new file, out.csv. So I dialed up each element, verified it, then did excel column copys which I then pasted into Cr6-Elements.

- Code:
If 21 == atomicNumber:
BuildProtonStack(2,i)
neutronGi.position = [0, 0, -pP1pP2/2 – p1n2] # (x,y,z)
slotset_df.iloc[i,8] = -pP1pP2/2 - p1n2 # Update the neutron’s z coord.
proStack5.add(neutronGi)
proStack5.position = [0, 0, -pP1pP2 - 4*orthoP]
slotset_df.iloc[i,3] = -pP1pP2 - 4*orthoP
grpBn.add(proStack5)
#slotlist[i] = 2
Above, is Scandium (Sc, 21) slot 5, copied from Atom Builder to mBuilder’s atom building section. One of 240 or so occurrences of AB’s 90 elements of up to 19 slots, all of which include the buildProtonStack() function. Altered in two ways; 1. Adding the proton z-coord (slotset_df.iloc[i,3])
and neutron z-coord (slotset_df.iloc[i,8]) values. The remaining x’s and y’s are equal to 0. And 2. Commenting out (#), the slotlist[] assignment, which is included in the data frame “Proton” column.
I clarified my neutron coding rules a bit. Such as:
1. For slots 1-15, Neutrons do not occupy proton-less slots, they must be positioned with respect to the adjacent proton occupied slot to which they are connected to.
2. For slots 16-19, Neutrons attached to the hook positions must occupy their own slots and are positioned with respect to those proton-less slots. Under the previous rule, protons in slots 2 and 3 might need to connect up to two pairs of neutrons, as I see it that would require several code changes. Just follow the two rules. The prospect of adding new neutrons is now easier.
The current plan is to build atoms from Cr6-Elements, including how to calculate new position sets as needed. No details yet, I may decide to include the calculation strings “-pP1pP2/2 – p1n2” and “-pP1pP2 – 4*orthoP” if I can figure out how to use them. I should read up on re - Regular expression operations https://docs.python.org/3/library/re.html
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
https://realpython.com/python-kwargs-and-args/
Chromium6- Posts : 826
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Perfect throw Cr6. I saw args and kwargs mentioned plenty of times, but since I didn’t see them in the pandas user guide or cookbook I thought they might be too esoteric. Thanks, I thoroughly enjoyed a totally new concept and appreciation of packing and unpacking functions. Then I reviewed another link or two and a couple of videos and forgot all about regex. * or **, allowing variable numbers of arguments or dictionaries passed into functions is a thing of beauty. An excellent solution for any number of problems. I haven’t thought of an applicable mBuilder one yet.
Most definite progress. All atoms are now built using Cr6-Elements.csv. All atomic slots’ protons as well as additional single neutrons or neutron pairs are properly assembled. Restructured the atom building section beginning on line 164. Total code lines for the atom building section was 1980 and is now down to 450 lines, many of them devoted to added neutrons.
- Code:
# Build each atom from Cr6-Elements
# atomicNumber = number # Select the desired atom from the dropdown widget or table.
grpAn = Group() # The rotation widget spins grpAn, slots 1,4 and 5.
grpBn = Group() # grpBn are the orthogonal top, bottom and hook slots.
if 1 <= number < 13:
caroSecs = 2.5
if 14 <= number < 20:
caroSecs = 4.0
if 21 <= number :
caroSecs = 6.0
proStack1 = Group()
proStack2 = Group()
proStack3 = Group()
proStack4 = Group()
proStack5 = Group()
proStack6 = Group()
proStack7 = Group()
proStack8 = Group()
proStack9 = Group()
proStack10 = Group()
proStack11 = Group()
proStack12 = Group()
proStack13 = Group()
proStack14 = Group()
proStack15 = Group()
proStack16 = Group()
proStack17 = Group()
proStack18 = Group()
proStack19 = Group()
# Dimensions # pRadius is now in In[1]
nRadius = pRadius*1.125 # Neutron radius
eRadius = 2*pRadius/10 # Electron radius
# Widget Controlled variables 1. ortho,
# 2.emR, 3. cAngleDeg, 4. pPTP, 5.p2p3Dist
pE = ortho*pRadius
emRadius = 2*emR*pRadius # 2*pE - pRadius/2
coneAngle = cAngleDeg*2*piG/360
coneHeight = emRadius*tan(coneAngle)
p12 = 2*pPTP*pRadius
p2p3 = 2*p2p3Dist*pRadius
n1n2 = (p12- 2*nRadius)/tan(coneAngle) # neutron-neutron orbital separation (y) inside the alpha
# e1e2 = -n1n2/10
p1p2 = pRadius*2.5 #0.75*p12 # pRadius*2.5
p1n1 = p12/2 #p12/2 #p12/2 # Alpha, up-down (z) proton-orbital neutron separation
p1n2 = p12/2 #pRadius + nRadius + 1.5*pRadius # In line proton and neutron
p1e1 = pRadius + 0.05*p12 # Alpha, top-bottom (z) proton-electron separation
maxZNeg = 0.0 # maxZNeg and maxZPos are Used to center(z) the atom on the screen
maxZPos = 0.0
# Proton set Geometries and Materials
protonGeometry = SphereBufferGeometry(pRadius, 24, 16)
neutronGeometry = SphereBufferGeometry(nRadius, 24, 16)
electronGeometry = SphereBufferGeometry(eRadius, 8, 16)
emissionGeometryP = CircleGeometry(emRadius, 24)
emissionGeometryX = ConeGeometry( emRadius, coneHeight, 16, openEnded = True )
protonMaterial = MeshStandardMaterial(color='blue')
protonMaterialB = MeshStandardMaterial(color='green')
neutronMaterial = MeshStandardMaterial(color='red')
neutronMaterialB = MeshStandardMaterial(color='green') # Color.js Find a nice color spectrum
electronMaterial = MeshStandardMaterial(color='cyan') #'orangered','salmon','green','cyan','teal','orange'
emissionmaterial7= MeshStandardMaterial(color='gray',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial1 = MeshStandardMaterial(color='magenta',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial2 = MeshStandardMaterial(color='blue',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial3 = MeshStandardMaterial(color='pink',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial4 = MeshStandardMaterial(color='salmon',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial5 = MeshStandardMaterial(color='lime',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial6 = MeshStandardMaterial(color='skyblue',side='DoubleSide',transparent = True,opacity = 0.20)
if emissionType.value == 'plane':
emissionGeometry = emissionGeometryP
if emissionType.value == 'cones':
emissionGeometry = emissionGeometryX
if coloredEmissions.value == True:
emissionmaterial = emissionmaterial1
else:
emissionmaterial = emissionmaterial7
def buildProtonStack(a,j): # a is the total # of protons. j indicates the calling register
""" The odd 'a' returns just the single, first, third, or fifth proton. The even 'a'
returns the first, second or third alpha set, each 2 protons and 2 neutrons. """
num = 0
if coloredEmissions.value == True:
if a == 1 : emissionmaterial = emissionmaterial1
if a == 2 : emissionmaterial = emissionmaterial2
if a == 3 : emissionmaterial = emissionmaterial3
if a == 4 : emissionmaterial = emissionmaterial4
if a == 5 : emissionmaterial = emissionmaterial5
if a == 6 : emissionmaterial = emissionmaterial6
else:
emissionmaterial = emissionmaterial7
# Proton z positions: # The possible formula solution, rather than the list, may be in these 2 lines
# protonMi.position = [0,0,a*p12/2+(a-1)*p2p3/2-(p2p3/2+p12/2)*num] # multiAlpha solution - Not
# protonMi.position = [0,0,(a//2)*p12/2+(a//2)*p2p3/2-(p2p3/2+p12/2)*num] # pSet
if a == 1 : pPositions = [0]
if a == 2 : pPositions = [p12/2,-p12/2]
if a == 3 : pPositions = [p12/2+p2p3/2,-p12/2+p2p3/2,-p12/2-p2p3/2]
if a == 4 : pPositions = [p12+p2p3/2,p2p3/2,-p2p3/2,-p12-p2p3/2]
if a == 5 : pPositions = [p12+p2p3,p2p3,0,-p12,-p12-p2p3]
if a == 6 : pPositions = [3*p12/2+p2p3,p12/2+p2p3,p12/2,-p12/2,-p12/2-p2p3,-3*p12/2-p2p3]
while num < a:
protonMi = Group()
protonMi.add(Mesh(protonGeometry, protonMaterial))
if emissionType.value == 'plane':
protonMi.add(Mesh(emissionGeometry, emissionmaterial))
if emissionType.value == 'cones':
emission1 = Object3D()
emission1.add(Mesh(emissionGeometry, emissionmaterial))
emission1.position = [0,0,coneHeight/2]
emission1.rotateX(-piG/2)
protonMi.add(emission1)
emission2 = Object3D()
emission2.add(Mesh(emissionGeometry, emissionmaterial))
emission2.position = [0,0,-coneHeight/2]
emission2.rotateX(piG/2)
protonMi.add(emission2)
protonMi.add(Mesh(emissionGeometryP, emissionmaterial))
electronMi = Object3D()
electronMi.add(Mesh(electronGeometry, electronMaterial))
electronMi.position = [0,-n1n2/10,-p1e1]
protonMi.add(electronMi)
protonMi.rotateZ(random()*2*piG)
if num%2 != 0:
neutronPair = Group()
neutronMi = Object3D()
neutronMi.add(Mesh(neutronGeometry, neutronMaterial))
neutronMi.position = [0,-n1n2/2,-p1n1]
neutronNi = Object3D()
neutronNi.add(Mesh(neutronGeometry, neutronMaterial))
neutronNi.position = [0,n1n2/2,-p1n1]
neutronPair.add(neutronMi)
neutronPair.add(neutronNi)
neutronPair.rotateZ(random()*2*piG)
protonMi.add(neutronPair)
protonMi.rotateX(piG)
protonMi.rotateZ(random()*2*piG)
protonMi.position = [0,0,pPositions[num]]
if j == 0 : proStack1.add(protonMi)
if j == 1 : proStack2.add(protonMi)
if j == 2 : proStack3.add(protonMi)
if j == 3 : proStack4.add(protonMi)
if j == 4 : proStack5.add(protonMi)
if j == 5 : proStack6.add(protonMi)
if j == 6 : proStack7.add(protonMi)
if j == 7 : proStack8.add(protonMi)
if j == 8 : proStack9.add(protonMi)
if j == 9 : proStack10.add(protonMi)
if j == 10 : proStack11.add(protonMi)
if j == 11 : proStack12.add(protonMi)
if j == 12 : proStack13.add(protonMi)
if j == 13 : proStack14.add(protonMi)
if j == 14 : proStack15.add(protonMi)
if j == 15 : proStack16.add(protonMi)
if j == 16 : proStack17.add(protonMi)
if j == 17 : proStack18.add(protonMi)
if j == 18 : proStack19.add(protonMi)
num += 1
return
# Assemble the atomic models.
for i in range( 19 ):
neutronGi = Object3D()
neutronGi.add(Mesh(neutronGeometry, neutronMaterial))
neutronKi = Object3D()
neutronKi.add(Mesh(neutronGeometry, neutronMaterial))
neutronPair = Group()
neutronLi = Object3D()
neutronLi.add(Mesh(neutronGeometry, neutronMaterial))
neutronLi.position = [0,-n1n2/2,0]
neutronPair.add(neutronLi)
neutronMi = Object3D()
neutronMi.add(Mesh(neutronGeometry, neutronMaterial))
neutronMi.position = [0,n1n2/2,0]
neutronPair.add(neutronMi)
neutronPair.rotateZ(random()*2*piG)
numPs = 0
numNs = 0
if (slotset_df.iloc[i,0] > 0) :
numPs = slotset_df.iloc[i,0]
if i == 0 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 : # Neutrons
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack1.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack1.add(neutronPair)
grpAn.add(proStack1)
if i == 1 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack2.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack2.add(neutronPair)
proStack2.rotateX(-piG/2)
locZ = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack2.position = [0, 0, locZ]
if slotset_df.iloc[15,9] > 0 : # Hook position neutrons
locN = p12
if slotset_df.iloc[15,9] == 1 :
neutronGi.position = [0,0,locN]
proStack2.add(neutronGi)
if slotset_df.iloc[15,9] == 2 :
neutronPair.position = [0,0,locN]
proStack2.add(neutronPair)
if slotset_df.iloc[17,9] > 0 :
locN = p12
if slotset_df.iloc[17,9] == 1 :
neutronGi.position = [0,0,-locN]
proStack2.add(neutronGi)
if slotset_df.iloc[17,9] == 2 :
neutronPair.position = [0,0,-locN]
proStack2.add(neutronPair)
grpBn.add(proStack2)
if i == 2 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack3.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack3.add(neutronPair)
proStack3.rotateX(-piG/2)
locZ = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack3.position = [0, 0, locZ]
if slotset_df.iloc[16,9] > 0 : # Hook position neutrons
locN = p12
if slotset_df.iloc[16,9] == 1 :
neutronGi.position = [0,0,-locN]
proStack3.add(neutronGi)
if slotset_df.iloc[16,9] == 2 :
neutronPair.position = [0,0,-locN]
proStack3.add(neutronPair)
if slotset_df.iloc[18,9] > 0 :
locN = p12
if slotset_df.iloc[18,9] == 1 :
neutronGi.position = [0,0,locN]
proStack3.add(neutronGi)
if slotset_df.iloc[18,9] == 2 :
neutronPair.position = [0,0,locN]
proStack3.add(neutronPair)
grpBn.add(proStack3)
if i == 3 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack4.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack4.add(neutronPair)
locZ = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack4.position = [0, 0, locZ]
grpBn.add(proStack4)
if i == 4 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack5.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack5.add(neutronPair)
locZ = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack5.position = [0, 0, locZ]
grpBn.add(proStack5)
if i == 5 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack6.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack6.add(neutronPair)
proStack6.rotateX(-piG/2)
locY = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack6.position = [0, locY, 0]
grpAn.add(proStack6)
if i == 6 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack7.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack7.add(neutronPair)
proStack7.rotateX(piG/2)
locY = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack7.position = [0, locY, 0]
grpAn.add(proStack7)
if i == 7 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack8.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack8.add(neutronPair)
proStack8.rotateX(-piG/2)
proStack8.rotateY(piG/2)
locX = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack8.position = [locX, 0, 0]
grpAn.add(proStack8)
if i == 8 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack9.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack9.add(neutronPair)
proStack9.rotateX(-piG/2)
proStack9.rotateY(piG/2)
locX = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack9.position = [locX, 0, 0]
grpAn.add(proStack9)
if i == 9 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack10.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack10.add(neutronPair)
locY = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack10.position = [0, locY, 0]
grpAn.add(proStack10)
if i == 10 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack11.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack11.add(neutronPair)
locX = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack11.position = [locX, 0, 0]
grpAn.add(proStack11)
if i == 11 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack12.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack12.add(neutronPair)
locY = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack12.position = [0, locY, 0]
grpAn.add(proStack12)
if i == 12 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack13.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack13.add(neutronPair)
locX = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack13.position = [locX, 0, 0]
grpAn.add(proStack13)
if i == 13 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack14.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack14.add(neutronPair)
proStack14.rotateX(-piG/2)
locZ = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack14.position = [0, 0, locZ]
grpBn.add(proStack14)
if i == 14 :
buildProtonStack(numPs,i)
if slotset_df.iloc[i,9] > 0 :
locN = slotset_df.iloc[i,13]*p12
if slotset_df.iloc[i,9] == 1 :
neutronGi.position = [0,0,locN]
proStack15.add(neutronGi)
if slotset_df.iloc[i,9] == 2 :
neutronPair.position = [0,0,locN]
proStack15.add(neutronPair)
proStack15.rotateX(-piG/2)
locZ = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack15.position = [0, 0, locZ]
grpBn.add(proStack15)
if i == 15 :
buildProtonStack(numPs,i)
locZ = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack16.position = [0, locZ, locZ]
grpBn.add(proStack16)
if i == 16 :
buildProtonStack(numPs,i)
locZ = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack17.position = [0, locZ, locZ]
grpBn.add(proStack17)
if i == 17 :
buildProtonStack(numPs,i)
locZ = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack18.position = [0, locZ, -locZ]
grpBn.add(proStack18)
if i == 18 :
buildProtonStack(numPs,i)
locZ = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack19.position = [0, locZ, -locZ]
grpBn.add(proStack19)
grpAn.position = [0,0,(-maxZNeg + maxZPos)/2 - maxZPos]
grpBn.position = [0,0,(-maxZNeg + maxZPos)/2 - maxZPos]
#print(valueL2z)
print(An) # (' 2 helium He ', 2)
print(slotlist)
#slotset_df.to_csv('out.csv')
slotset_df
The crucial details didn’t occur to me until after my last post, when I reported adding all occupied slot default location values in columns “PX”, “PY”, and “PZ”. I realized what the data needed was building instructions which just so happened to be available in each slot location calculation, the location control variables.
So I simplified a couple of the control variable ‘labels’, changing orthoP to pE, and changing pP1pP2 to p12, making all the coded locations from AB easier to read. Then added 4 new columns for the 3 variables and an unused summation after column "PZ": 1. "P2P3", 2 "P12", 3. "PE", and 4. "PSCalc". All occupied slot locations are determined by the summation of the products of those variables and that slot’s particular coefficients. Each proton occupied slot now includes those coefficients.
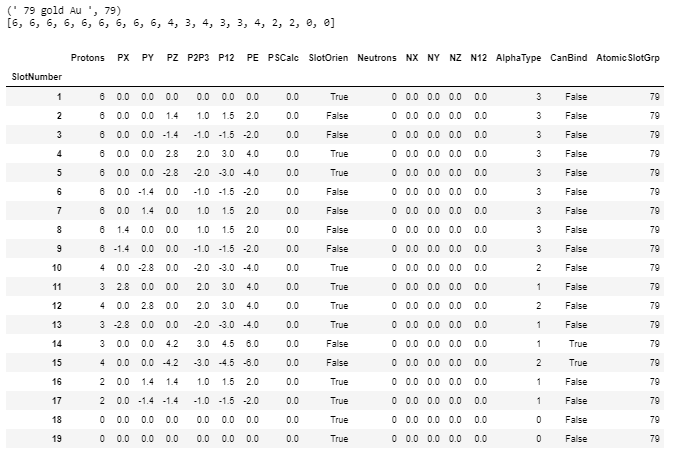
Selecting an atom also selects that atom’s 19 row dataframe - slotset_df. Now, upon execution, mbuilder’s atom building section reads and builds each occupied slot row of slotset_df. Having changed the control variables or not, all variables are ‘fixed’ and all positions are recalculated. The default values give a sense of the magnitude and direction of each atomic position, but “PX”, “PY”, and “PZ” aren’t currently ‘unused’.
Neutrons. Added new column after "NZ": “N12”. n1n2 = p12/2, the z distance between a neutron and the center of the slot in terms of p12. The neutron is then rotated and re positioned as needed. All neutrons are now attached to proton occupied slots including neutrons attached to the hook positions. Another humble lesson, when I gave the neutrons their own slots they spun in the wrong orientation – quickly rejected that idea. Seems a bit cumbersome, notice how slot 2 (196-225) or slot 3 (226-256) handle those cases where extra neutrons appear in the hook positions (slots 16-19). I might be able to eliminate a couple of lines. I’m certainly still want to add more neutrons.
I’ll try adding another tab for slotset_df. What about these unused columns? Plenty of cleanup needed. For example, maxZPos and maxZNeg aren’t used. I may wander around a bit.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
LloydK- Posts : 548
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
The atom building section.
- Code:
# Build each atom from Cr6-Elements
# atomicNumber = number # Select the desired atom from the dropdown widget or table.
grpAn = Group() # The rotation widget spins grpAn, slots 1,4 and 5.
grpBn = Group() # grpBn are the orthogonal top, bottom and hook slots.
if 1 <= number < 13:
caroSecs = 2.5
if 14 <= number < 20:
caroSecs = 4.0
if 21 <= number :
caroSecs = 6.0
proStack1 = Group()
proStack2 = Group()
proStack3 = Group()
proStack4 = Group()
proStack5 = Group()
proStack6 = Group()
proStack7 = Group()
proStack8 = Group()
proStack9 = Group()
proStack10 = Group()
proStack11 = Group()
proStack12 = Group()
proStack13 = Group()
proStack14 = Group()
proStack15 = Group()
proStack16 = Group()
proStack17 = Group()
proStack18 = Group()
proStack19 = Group()
# Dimensions # pRadius is now in In[1]
nRadius = pRadius*1.125 # Neutron radius
eRadius = 2*pRadius/10 # Electron radius
# Widget Controlled variables 1. ortho,
# 2.emR, 3. cAngleDeg, 4. pPTP, 5.p2p3Dist
pE = ortho*pRadius
emRadius = 2*emR*pRadius # 2*pE - pRadius/2
coneAngle = cAngleDeg*2*piG/360
coneHeight = emRadius*tan(coneAngle)
p12 = 2*pPTP*pRadius
p2p3 = 2*p2p3Dist*pRadius
n1n2 = (p12- 2*nRadius)/tan(coneAngle) # neutron-neutron orbital separation (y) inside the alpha
# e1e2 = -n1n2/10
p1p2 = pRadius*2.5 #0.75*p12 # pRadius*2.5
p1n1 = p12/2 #p12/2 #p12/2 # Alpha, up-down (z) proton-orbital neutron separation
p1n2 = p12/2 #pRadius + nRadius + 1.5*pRadius # In line proton and neutron
p1e1 = pRadius + 0.05*p12 # Alpha, top-bottom (z) proton-electron separation
#maxZNeg = 0.0 # maxZNeg and maxZPos are Used to center(z) the atom on the screen
#maxZPos = 0.0
# Proton set Geometries and Materials
protonGeometry = SphereBufferGeometry(pRadius, 24, 16)
neutronGeometry = SphereBufferGeometry(nRadius, 24, 16)
electronGeometry = SphereBufferGeometry(eRadius, 8, 16)
emissionGeometryP = CircleGeometry(emRadius, 24)
emissionGeometryX = ConeGeometry( emRadius, coneHeight, 16, openEnded = True )
protonMaterial = MeshStandardMaterial(color='blue')
protonMaterialB = MeshStandardMaterial(color='green')
neutronMaterial = MeshStandardMaterial(color='red')
neutronMaterialB = MeshStandardMaterial(color='green') # Color.js Find a nice color spectrum
electronMaterial = MeshStandardMaterial(color='cyan') #'orangered','salmon','green','cyan','teal','orange'
emissionmaterial7= MeshStandardMaterial(color='gray',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial1 = MeshStandardMaterial(color='magenta',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial2 = MeshStandardMaterial(color='blue',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial3 = MeshStandardMaterial(color='pink',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial4 = MeshStandardMaterial(color='salmon',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial5 = MeshStandardMaterial(color='lime',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial6 = MeshStandardMaterial(color='skyblue',side='DoubleSide',transparent = True,opacity = 0.20)
if emissionType.value == 'plane':
emissionGeometry = emissionGeometryP
if emissionType.value == 'cones':
emissionGeometry = emissionGeometryX
if coloredEmissions.value == True:
emissionmaterial = emissionmaterial1
else:
emissionmaterial = emissionmaterial7
def buildProtonStack(a,j): # a is the total # of protons. j indicates the calling register
""" The odd 'a' returns just the single, first, third, or fifth proton. The even 'a'
returns the first, second or third alpha set, each 2 protons and 2 neutrons. """
num = 0
if coloredEmissions.value == True:
if a == 1 : emissionmaterial = emissionmaterial1
if a == 2 : emissionmaterial = emissionmaterial2
if a == 3 : emissionmaterial = emissionmaterial3
if a == 4 : emissionmaterial = emissionmaterial4
if a == 5 : emissionmaterial = emissionmaterial5
if a == 6 : emissionmaterial = emissionmaterial6
else:
emissionmaterial = emissionmaterial7
# Proton z positions: # The possible formula solution, rather than the list, may be in these 2 lines
# protonMi.position = [0,0,a*p12/2+(a-1)*p2p3/2-(p2p3/2+p12/2)*num] # multiAlpha solution - Not
# protonMi.position = [0,0,(a//2)*p12/2+(a//2)*p2p3/2-(p2p3/2+p12/2)*num] # pSet
if a == 1 : pPositions = [0]
if a == 2 : pPositions = [p12/2,-p12/2]
if a == 3 : pPositions = [p12/2+p2p3/2,-p12/2+p2p3/2,-p12/2-p2p3/2]
if a == 4 : pPositions = [p12+p2p3/2,p2p3/2,-p2p3/2,-p12-p2p3/2]
if a == 5 : pPositions = [p12+p2p3,p2p3,0,-p12,-p12-p2p3]
if a == 6 : pPositions = [3*p12/2+p2p3,p12/2+p2p3,p12/2,-p12/2,-p12/2-p2p3,-3*p12/2-p2p3]
while num < a:
protonMi = Group()
protonMi.add(Mesh(protonGeometry, protonMaterial))
if emissionType.value == 'plane':
protonMi.add(Mesh(emissionGeometry, emissionmaterial))
if emissionType.value == 'cones':
emission1 = Object3D()
emission1.add(Mesh(emissionGeometry, emissionmaterial))
emission1.position = [0,0,coneHeight/2]
emission1.rotateX(-piG/2)
protonMi.add(emission1)
emission2 = Object3D()
emission2.add(Mesh(emissionGeometry, emissionmaterial))
emission2.position = [0,0,-coneHeight/2]
emission2.rotateX(piG/2)
protonMi.add(emission2)
protonMi.add(Mesh(emissionGeometryP, emissionmaterial))
electronMi = Object3D()
electronMi.add(Mesh(electronGeometry, electronMaterial))
electronMi.position = [0,-n1n2/10,-p1e1]
protonMi.add(electronMi)
protonMi.rotateZ(random()*2*piG)
if num%2 != 0:
neutronPair = Group()
neutronMi = Object3D()
neutronMi.add(Mesh(neutronGeometry, neutronMaterial))
neutronMi.position = [0,-n1n2/2,-p1n1]
neutronNi = Object3D()
neutronNi.add(Mesh(neutronGeometry, neutronMaterial))
neutronNi.position = [0,n1n2/2,-p1n1]
neutronPair.add(neutronMi)
neutronPair.add(neutronNi)
neutronPair.rotateZ(random()*2*piG)
protonMi.add(neutronPair)
protonMi.rotateX(piG)
protonMi.rotateZ(random()*2*piG)
protonMi.position = [0,0,pPositions[num]]
if j == 0 : proStack1.add(protonMi)
if j == 1 : proStack2.add(protonMi)
if j == 2 : proStack3.add(protonMi)
if j == 3 : proStack4.add(protonMi)
if j == 4 : proStack5.add(protonMi)
if j == 5 : proStack6.add(protonMi)
if j == 6 : proStack7.add(protonMi)
if j == 7 : proStack8.add(protonMi)
if j == 8 : proStack9.add(protonMi)
if j == 9 : proStack10.add(protonMi)
if j == 10 : proStack11.add(protonMi)
if j == 11 : proStack12.add(protonMi)
if j == 12 : proStack13.add(protonMi)
if j == 13 : proStack14.add(protonMi)
if j == 14 : proStack15.add(protonMi)
if j == 15 : proStack16.add(protonMi)
if j == 16 : proStack17.add(protonMi)
if j == 17 : proStack18.add(protonMi)
if j == 18 : proStack19.add(protonMi)
num += 1
return
def Orient(n, stack):
# The proton stack emission plane for proStacks are oriented in the z-plane.
# Reorient the stack otherwise according to the "slotOrien" column.
if slotset_df.loc[n+1,('SlotOrien')] == "Y" :
stack.rotateX(-piG/2)
if slotset_df.loc[n+1,('SlotOrien')] == "X" :
stack.rotateX(-piG/2)
stack.rotateY(piG/2)
return
def Position(n,stack):
# Calculate the slot's position based on the the current values
# of p2p3,p12 and pE. The default vales are just position markers.
loc = abs(slotset_df.loc[n+1,('P2P3')]*p2p3 + slotset_df.loc[n+1,('P12')]*p12 + slotset_df.loc[n+1,('PE')]*pE)
# Position the stack according to the presence of slotset's
# default values (make 'em one) and sign.
px = slotset_df.loc[i+1,('PX')]
py = slotset_df.loc[i+1,('PY')]
pz = slotset_df.loc[i+1,('PZ')]
if px != 0 : px = copysign(1, px)
if py != 0 : py = copysign(1, py)
if pz != 0 : pz = copysign(1, pz)
stack.position = [px*loc, py*loc, pz*loc]
return
# Assemble the atomic models.
for i in range( 19 ):
neutronGi = Object3D()
neutronGi.add(Mesh(neutronGeometry, neutronMaterial))
neutronKi = Object3D()
neutronKi.add(Mesh(neutronGeometry, neutronMaterial))
neutronPair = Group()
neutronLi = Object3D()
neutronLi.add(Mesh(neutronGeometry, neutronMaterial))
neutronLi.position = [0,-n1n2/2,0]
neutronPair.add(neutronLi)
neutronMi = Object3D()
neutronMi.add(Mesh(neutronGeometry, neutronMaterial))
neutronMi.position = [0,n1n2/2,0]
neutronPair.add(neutronMi)
neutronPair.rotateZ(random()*2*piG)
numPs = 0
numNs = 0
if slotset_df.loc[i+1,('Protons')] > 0 :
numPs = slotset_df.loc[i+1,('Protons')]
if i == 0 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 : # Neutrons
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack1.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack1.add(neutronPair)
# Orien Rotates the proton stack according to slotset_df col 'SlotOrien'
Orient(i,proStack1)
Position(i,proStack1) # It's (0,0,0)
grpAn.add(proStack1)
if i == 1 :
buildProtonStack(numPs,i)
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[16,('Neutrons')] > 0 : # Hook position neutrons
locN = p12
if slotset_df.loc[16,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack2.add(neutronGi)
if slotset_df.loc[16,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack2.add(neutronPair)
if slotset_df.loc[18,('Neutrons')] > 0 :
locN = p12
if slotset_df.loc[18,('Neutrons')] == 1 :
neutronGi.position = [0,0,-locN]
proStack2.add(neutronGi)
if slotset_df.loc[18,('Neutrons')] == 2 :
neutronPair.position = [0,0,-locN]
proStack2.add(neutronPair)
Orient(i,proStack2)
Position(i,proStack2)
grpBn.add(proStack2)
if i == 2 :
buildProtonStack(numPs,i)
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[17,('Neutrons')] > 0 : # Hook position neutrons
locN = p12
if slotset_df.loc[17,('Neutrons')] == 1 :
neutronGi.position = [0,0,-locN]
proStack3.add(neutronGi)
if slotset_df.loc[17,('Neutrons')] == 2 :
neutronPair.position = [0,0,-locN]
proStack3.add(neutronPair)
if slotset_df.loc[19,('Neutrons')] > 0 :
locN = p12
if slotset_df.loc[19,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack3.add(neutronGi)
if slotset_df.loc[19,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack3.add(neutronPair)
Orient(i,proStack3)
Position(i,proStack3)
grpBn.add(proStack3)
if i == 3 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack4.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack4.add(neutronPair)
Orient(i,proStack4)
Position(i,proStack4)
grpBn.add(proStack4)
if i == 4 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack5.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack5.add(neutronPair)
#locZ = slotset_df.loc[i+1,('P2P3')]*p2p3 + slotset_df.loc[i+1,('P12')]*p12 + slotset_df.loc[i+1,('PE')]*pE
Orient(i,proStack5)
Position(i,proStack5)
#proStack5.position = [0, 0, locZ]
grpBn.add(proStack5)
if i == 5 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack6.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack6.add(neutronPair)
Orient(i,proStack6)
Position(i,proStack6)
grpAn.add(proStack6)
if i == 6 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack7.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack7.add(neutronPair)
Orient(i,proStack7)
Position(i,proStack7)
grpAn.add(proStack7)
if i == 7 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack8.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack8.add(neutronPair)
Orient(i,proStack8)
Position(i,proStack8)
grpAn.add(proStack8)
if i == 8 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack9.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack9.add(neutronPair)
Orient(i,proStack9)
Position(i,proStack9)
grpAn.add(proStack9)
if i == 9 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack10.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack10.add(neutronPair)
Orient(i,proStack10)
Position(i,proStack10)
grpAn.add(proStack10)
if i == 10 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack11.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack11.add(neutronPair)
Orient(i,proStack11)
Position(i,proStack11)
grpAn.add(proStack11)
if i == 11 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack12.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack12.add(neutronPair)
Orient(i,proStack12)
Position(i,proStack12)
grpAn.add(proStack12)
if i == 12 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack13.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack13.add(neutronPair)
Orient(i,proStack13)
Position(i,proStack13)
grpAn.add(proStack13)
if i == 13 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack14.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack14.add(neutronPair)
Orient(i,proStack14)
Position(i,proStack14)
grpBn.add(proStack14)
if i == 14 :
buildProtonStack(numPs,i)
if slotset_df.loc[i+1,('Neutrons')] > 0 :
locN = slotset_df.loc[i+1,('N12')]*p12
if slotset_df.loc[i+1,('Neutrons')] == 1 :
neutronGi.position = [0,0,locN]
proStack15.add(neutronGi)
if slotset_df.loc[i+1,('Neutrons')] == 2 :
neutronPair.position = [0,0,locN]
proStack15.add(neutronPair)
Orient(i,proStack15)
Position(i,proStack15)
grpBn.add(proStack15)
if i == 15 :
# Slots 16-19 are shorter - no added neutrons
buildProtonStack(numPs,i)
Orient(i,proStack16)
Position(i,proStack16)
grpBn.add(proStack16)
if i == 16 :
buildProtonStack(numPs,i)
Orient(i,proStack17)
Position(i,proStack17)
grpBn.add(proStack17)
if i == 17 :
buildProtonStack(numPs,i)
Orient(i,proStack18)
Position(i,proStack18)
grpBn.add(proStack18)
if i == 18 :
buildProtonStack(numPs,i)
Orient(i,proStack19)
Position(i,proStack19)
grpBn.add(proStack19)
print(An) # (' 2 helium He ', 2)
print(slotlist)
#slotset_df.to_csv('out.csv')
slotset_df
Changes since last time:
1. Got rid of unused maxZPos and maxZNeg used to center the atom in the z dimension. They may come back when atoms need to be added together.
2. The slot rotation controls were still in the Atom Builder (AB) slotlayout (SL) configuration. I followed my instructions above, starting with: switch 6 and 14, etc, to reconfigure the rotation button-actions to match mBuilder’s SL diagram.
3. "SlotOrien" column, H or V values - Wrong. In our charge field model, proton and proton stack emission planes are oriented in either the X plane (slots 8,9), Y plane (slots 2,3,6,7,14,15), or Z plane (slots 1,4,5,10,11,12,13,16,17,18,19). Made that column values content change to each row, all 90 lists of 19 slot orientations are the same.
4. The code I posted last time included details which were not yet read from the .csv file. Example.
proStack15.rotateX(-piG/2)
It worked but there was no explanation why proStack15 was given a negative 90 degree x-rotation, its built into the assembly code. I replaced that line with:
Orient(i,proStack15)
after having created an Orient() function which rotates each stack, or not , according to the "SlotOrien" column’s X, Y or Z values.
- Code:
def Orient(n, stack):
# The proton stack emission plane for proStacks are oriented in the z-plane.
# Reorient the stack otherwise according to the "slotOrien" column.
if slotset_df.loc[n+1,('SlotOrien')] == "Y" :
stack.rotateX(-piG/2)
if slotset_df.loc[n+1,('SlotOrien')] == "X" :
stack.rotateX(-piG/2)
stack.rotateY(piG/2)
return
5. Last week’s code used what’s known as integer based indexing; dot iloc square brackets, .iloc[:,:]. The pandas.DataFrame.iloc property. The square brackets contain [row,col] indexes. For example,
locY = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
Changed all .iloc[] to label based - loc[] indexing. The labels are the column names, making all the code easier to read. Here’s that same line with .loc[], changing from (col) 4 to 'P2P3', from (col) 5 to ‘P12’, and from (col) 6 to ‘PE’.
loc = abs(slotset_df.loc[n+1,('P2P3')]*p2p3 + slotset_df.loc[n+1,('P12')]*p12 + slotset_df.loc[n+1,('PE')]*pE)
Sorry, I just noticed the confusing/misleading variable name loc - short for location, or locN, locX, locY or locZ in various places. Those are variable names for calculated positions and are not indexes.
I tried to do .loc[] indexing from the start, but the code didn’t cooperate and so I went with iloc instead. Turns out that .iloc’s i = .loc’s i+1. I believe that’ may be due to the fact that slotset_df’s ‘labeled’ row index is numbered according to the slot numbers 1-19 instead of the integer index default 0-18.
6. Another detail hidden in the code was where to put the proton stack => +/-X,+/-Y or +/-Z, for slots 2-15, (slot 1 is at (0,0,0); then there are both +/-Y and +/-Z coordinates for the hook positions, slots 16-19. Here’s an example where the y coordinate is specified, and not read from the data file.
locY = slotset_df.iloc[i,4]*p2p3 + slotset_df.iloc[i,5]*p12 + slotset_df.iloc[i,6]*pE
proStack12.position = [0, locY, 0]
Changed that code to:
Position(i,proStack15)
After having created a Position() function which determines the stack’s position/direction (from (0,0,0)) according to the column positions and signs from the table’s default values.
- Code:
def Position(n,stack):
# Calculate the slot's position based on the current values
# of p2p3, p12 and pE. The default values are just position markers.
loc = abs(slotset_df.loc[n+1,('P2P3')]*p2p3 + slotset_df.loc[n+1,('P12')]*p12 + slotset_df.loc[n+1,('PE')]*pE)
# Position the stack according to the presence of slotset's
# default values (make 'em one) and sign.
px = slotset_df.loc[i+1,('PX')]
py = slotset_df.loc[i+1,('PY')]
pz = slotset_df.loc[i+1,('PZ')]
if px != 0 : px = copysign(1, px)
if py != 0 : py = copysign(1, py)
if pz != 0 : pz = copysign(1, pz)
stack.position = [px*loc, py*loc, pz*loc]
return
stack.position = [px*loc, py*loc, pz*loc]
One always calls a function with its name, followed by parentheses which contain the passed parameters as I did above with (n, stack). n is the slot number, and stack works the passed proStack.
7. Updated mBuilder’s first cell with: a new update, introductory text and added operating instructions. Also added the operating instructions to the “Atom data” cell, telling the user to start the notebook from there.
/\\///\\\\/////\\\\\\/////\\\\///\\/
Wasn’t able to add slotset_df to an ipywidgit tab. Starting with these two stackoverflow questions.
https://stackoverflow.com/questions/55351565/display-pandas-dataframe-into-another-tab
https://stackoverflow.com/questions/39977117/ipython-display-full-dataframe-in-new-tab?noredirect=1&lq=1
Won’t pursue it. The slotset_df output works fine where it is, printed right after the atom building cell.
\//\\\////\\\\\//////\\\\\////\\\//\
Neutrons. Please pardon my broken record neutron mentions, forget all my nonsense about neutrons appearing in the slot positions or not. I intend to make this work right eventually.
I guess it depends on the atom’s atomic weight; but I assume there should be roughly equal numbers of protons and neutrons for a given atom. It has nothing to do with parity or charge balance, larger atoms offer plenty of safe havens for neutrons. I imagine it would take plenty of time, energy and effort to completely pack an atom with neutrons. Anyway, the program adds two neutrons for every alpha in every atom; plus 47 neutrons, mostly added to “twenties” atomic numbers, from scandium (21) through copper (29). Copper is a one-of-a-kind special case example with not only one, but two pairs of neutrons in two slot positions. Those neutron additions were made according to images in the original Cr6-Elements file, from Miles’ own diagrams. I haven’t made any other neutron additions of my own.
Running the numbers, across all 90 atoms, there are 4095 protons (90+89+88+…) and 3687 neutrons (including the 47 additions mentioned). Elsewhere, the Periodic table neutron deficit is over 400.
I attempted to start adding neutrons to the atoms with the largest neutron deficits (between 10 to 16 for each atom), the lanthanide series, 57-70, all containing protonstacks with 5 protons for which the buildProStack() function adds only 4 neutrons. I was soon stymied. I’ll probably need to change buildProStack() too.
Putting my thoughts together, consider the following. Let the main charge flow be from left to right. Let a vertical pipe character (|) represent a proton with vertical edgewise emission, so the proton’s poles are aligned to the main L=>R charge flow direction. Let one neutron be represented by a period(.) and a pair of neutrons with a colon(
- Code:
Hiding from Smiley Face changes to my proton/neutron notation.
Here’s a bare proton, no neutrons, a. [ | ].
Hydrogen with a single neutron on the left and right sides of the proton: b. [ .| ], c. [ |. ].
Deuterium. Two neutrons: d. [ :| ], e. [ .|. ], f. [ |: ].
Tritium. Three neutrons: g. [ :.| ]; h. [ :|. ]; i. [ .|: ]; j. [ |.: ].
The program doesn’t include any neutrons on both sides of a proton stack (examples e., h., i.) nor any sets of three neutrons, but I’d like to accommodate any of those 10 options - in keeping with the charge flow direction of course. Without adding any new rows. Maybe adding a neutron configuration column indicating that slot’s a through j, or one of ten possible neutron configurations for a slot with zero to 3 neutrons.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
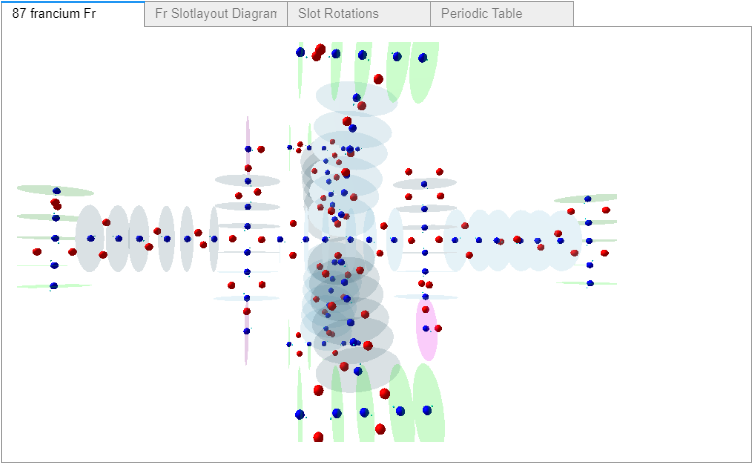
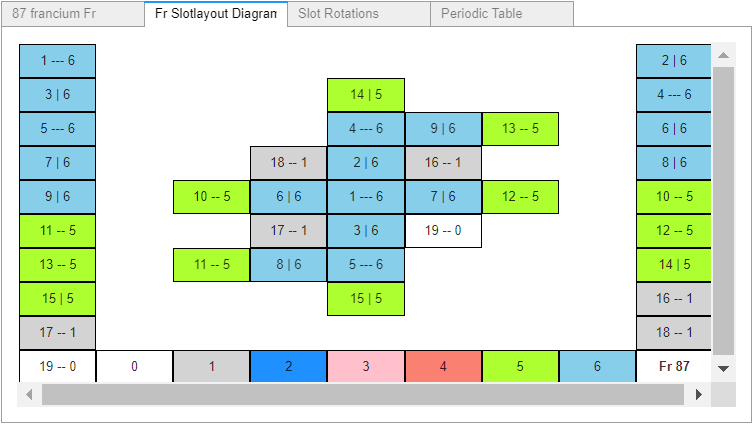
This week’s 90 element proton/neutron totals: Tot. protons = 4095. Tot. neutrons = 4055. Alpha Ns = 3640. Added Ns = 415; minus last week’s 47 makes 368 neutrons added since last week. Along with the necessary code changes and additions to Cr6-Elements.csv.
I must say, these neutron additions show HuAhh class progress. Sure there are questions on exactly where and under what conditions additional neutrons may be present, still, the code shift to the datafile makes it far easier to make subsequent changes; such as possible variable or random neutron counts.
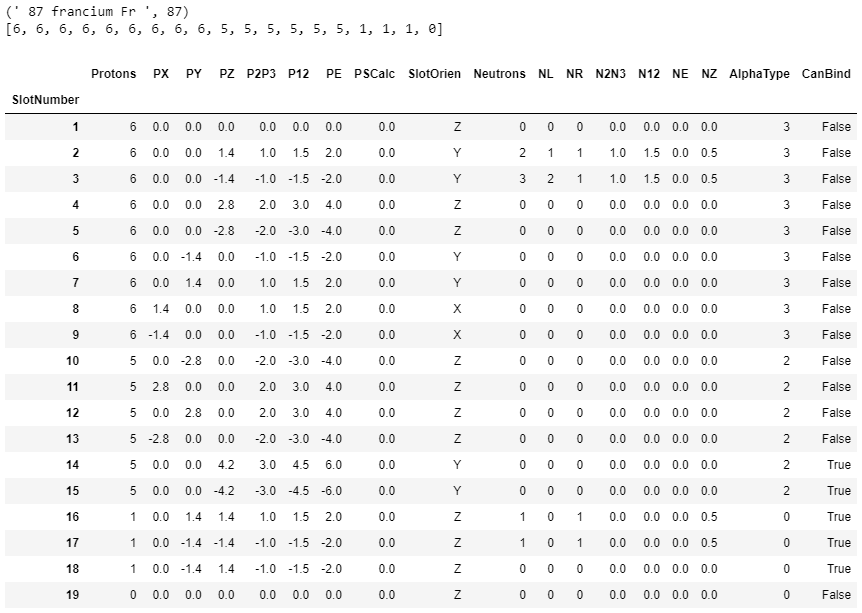
The improved buildProtonStackN(a,j) function now adds all alpha and non-alpha neutrons according to each element’s slotset_df. Each slot’s left and/or right ends, columns “NL” and “NR”, can now have 0-2 neutrons added. A third additional L and/or R neutron hasn't been coded yet but it can fit right in. The neutron positions, like the proton’s, depend on user controlled variables p12, the distance between the two protons in a single alpha; and p2p3, the pole-to-pole distance between alphas. pE, the male-proton/female-proton distance between adjacent proton emission and proton pole is outside the stack and doesn’t currently effect neutron positions.
- Code:
# Build each atom from Cr6-Elements
# atomicNumber = number # Select the desired atom from the dropdown widget or table.
grpAn = Group() # The rotation widget spins grpAn, slots 1,4 and 5.
grpBn = Group() # grpBn are the orthogonal top, bottom and hook slots.
if 1 <= number < 13:
caroSecs = 2.5
if 14 <= number < 20:
caroSecs = 4.0
if 21 <= number :
caroSecs = 6.0
proStack1 = Group()
proStack2 = Group()
proStack3 = Group()
proStack4 = Group()
proStack5 = Group()
proStack6 = Group()
proStack7 = Group()
proStack8 = Group()
proStack9 = Group()
proStack10 = Group()
proStack11 = Group()
proStack12 = Group()
proStack13 = Group()
proStack14 = Group()
proStack15 = Group()
proStack16 = Group()
proStack17 = Group()
proStack18 = Group()
proStack19 = Group()
# Dimensions # pRadius is now in In[1]
nRadius = pRadius*1.125 # Neutron radius
eRadius = 2*pRadius/10 # Electron radius
# Widget Controlled variables 1. ortho,
# 2.emR, 3. cAngleDeg, 4. pPTP, 5.p2p3Dist
pE = ortho*pRadius
emRadius = 2*emR*pRadius # 2*pE - pRadius/2
coneAngle = cAngleDeg*2*piG/360
coneHeight = emRadius*tan(coneAngle)
p12 = 2*pPTP*pRadius
p2p3 = 2*p2p3Dist*pRadius
n1n2 = (p12- 2*nRadius)/tan(coneAngle) # neutron-neutron orbital separation (y) inside the alpha
# e1e2 = -n1n2/10
p1p2 = pRadius*2.5 #0.75*p12 # pRadius*2.5
p1n1 = p12/2 #p12/2 #p12/2 # Alpha, up-down (z) proton-orbital neutron separation
p1n2 = p12/2 #pRadius + nRadius + 1.5*pRadius # In line proton and neutron
p1e1 = pRadius + 0.05*p12 # Alpha, top-bottom (z) proton-electron separation
#maxZNeg = 0.0 # maxZNeg and maxZPos are Used to center(z) the atom on the screen
#maxZPos = 0.0
# Proton set Geometries and Materials
protonGeometry = SphereBufferGeometry(pRadius, 24, 16)
neutronGeometry = SphereBufferGeometry(nRadius, 24, 16)
electronGeometry = SphereBufferGeometry(eRadius, 8, 16)
emissionGeometryP = CircleGeometry(emRadius, 24)
emissionGeometryX = ConeGeometry( emRadius, coneHeight, 16, openEnded = True )
protonMaterial = MeshStandardMaterial(color='blue')
protonMaterialB = MeshStandardMaterial(color='green')
neutronMaterial = MeshStandardMaterial(color='red')
neutronMaterialB = MeshStandardMaterial(color='green') # Color.js Find a nice color spectrum
electronMaterial = MeshStandardMaterial(color='cyan') #'orangered','salmon','green','cyan','teal','orange'
emissionmaterial7= MeshStandardMaterial(color='gray',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial1 = MeshStandardMaterial(color='magenta',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial2 = MeshStandardMaterial(color='blue',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial3 = MeshStandardMaterial(color='pink',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial4 = MeshStandardMaterial(color='salmon',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial5 = MeshStandardMaterial(color='lime',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial6 = MeshStandardMaterial(color='skyblue',side='DoubleSide',transparent = True,opacity = 0.20)
if emissionType.value == 'plane':
emissionGeometry = emissionGeometryP
if emissionType.value == 'cones':
emissionGeometry = emissionGeometryX
if coloredEmissions.value == True:
emissionmaterial = emissionmaterial1
else:
emissionmaterial = emissionmaterial7
def buildProtonStackN(a,j):
# a is the total number of protons. j indicates the calling slot #.
# Each of the "a" protons in a given stack is added one at a time;
# either as the first "odd" proton added alone - plus electron that
# is, or as the second "even" proton which is added along with two
# neutrons in order to create an alpha.
num = 0
if coloredEmissions.value == True:
if a == 1 : emissionmaterial = emissionmaterial1
if a == 2 : emissionmaterial = emissionmaterial2
if a == 3 : emissionmaterial = emissionmaterial3
if a == 4 : emissionmaterial = emissionmaterial4
if a == 5 : emissionmaterial = emissionmaterial5
if a == 6 : emissionmaterial = emissionmaterial6
else:
emissionmaterial = emissionmaterial7
# Proton z positions: # The possible formula solution, rather than the list, may be in these 2 lines
# protonMi.position = [0,0,a*p12/2+(a-1)*p2p3/2-(p2p3/2+p12/2)*num] # multiAlpha solution - Not
# protonMi.position = [0,0,(a//2)*p12/2+(a//2)*p2p3/2-(p2p3/2+p12/2)*num] # pSet
if a == 1 : pPositions = [0]
if a == 2 : pPositions = [p12/2,-p12/2]
if a == 3 : pPositions = [p12/2+p2p3/2,-p12/2+p2p3/2,-p12/2-p2p3/2]
if a == 4 : pPositions = [p12+p2p3/2,p2p3/2,-p2p3/2,-p12-p2p3/2]
if a == 5 : pPositions = [p12+p2p3,p2p3,0,-p12,-p12-p2p3]
if a == 6 : pPositions = [3*p12/2+p2p3,p12/2+p2p3,p12/2,-p12/2,-p12/2-p2p3,-3*p12/2-p2p3]
ncounter = 0
rcounter = 0
lcounter = 0
while num < a:
protonMi = Group()
protonMi.add(Mesh(protonGeometry, protonMaterial))
if emissionType.value == 'plane':
protonMi.add(Mesh(emissionGeometry, emissionmaterial))
if emissionType.value == 'cones':
emission1 = Object3D()
emission1.add(Mesh(emissionGeometry, emissionmaterial))
emission1.position = [0,0,coneHeight/2]
emission1.rotateX(-piG/2)
protonMi.add(emission1)
emission2 = Object3D()
emission2.add(Mesh(emissionGeometry, emissionmaterial))
emission2.position = [0,0,-coneHeight/2]
emission2.rotateX(piG/2)
protonMi.add(emission2)
protonMi.add(Mesh(emissionGeometryP, emissionmaterial))
electronMi = Object3D()
electronMi.add(Mesh(electronGeometry, electronMaterial))
electronMi.position = [0,-n1n2/10,-p1e1]
protonMi.add(electronMi)
protonMi.rotateZ(random()*2*piG)
if num%2 != 0:
neutronPair = Group()
neutronMi = Object3D()
neutronMi.add(Mesh(neutronGeometry, neutronMaterial))
neutronMi.position = [0,-n1n2/2,-p1n1]
neutronNi = Object3D()
neutronNi.add(Mesh(neutronGeometry, neutronMaterial))
neutronNi.position = [0,n1n2/2,-p1n1]
neutronPair.add(neutronMi)
neutronPair.add(neutronNi)
neutronPair.rotateZ(random()*2*piG)
protonMi.add(neutronPair)
protonMi.rotateX(piG)
protonMi.rotateZ(random()*2*piG)
# Neutron Additions. Add neutrons where specified by the data file.
# This code adds any and all left or right neutrons during the
# addition of the "a" sized stack's first proton. pPositions[0]
# is the center for all neutrons added to that stack. Subtracting
# pPositions[0] brings the neutrons center to the center of the
# stack, so the neutron placement calculation applies equally for
# both left and right ends.
if (slotset_df.loc[i+1,('NL')] > 0) and (lcounter == 0) :
locN = slotset_df.loc[i+1,('N2N3')]*p2p3 + slotset_df.loc[i+1,('N12')]*p12 + slotset_df.loc[i+1,('NZ')]*p12 - pPositions[0]
if slotset_df.loc[i+1,('NL')] == 1 :
neutronVi = Object3D()
neutronVi.add(Mesh(neutronGeometry, neutronMaterial))
neutronVi.position = [0,0,locN]
protonMi.add(neutronVi)
lcounter += 1
if slotset_df.loc[i+1,('NL')] == 2 :
neutronGi = Object3D()
neutronGi.add(Mesh(neutronGeometry, neutronMaterial))
neutronKi = Object3D()
neutronKi.add(Mesh(neutronGeometry, neutronMaterial))
neutronPair = Group()
neutronGi.position = [0,-n1n2/2,0]
neutronPair.add(neutronGi)
neutronKi.position = [0,n1n2/2,0]
neutronPair.add(neutronKi)
neutronPair.rotateZ(random()*2*piG)
neutronPair.position = [0,0,locN]
protonMi.add(neutronPair)
lcounter += 1
if (slotset_df.loc[i+1,('NR')] > 0) and (rcounter == 0) :
locN = - slotset_df.loc[i+1,('N2N3')]*p2p3 - slotset_df.loc[i+1,('N12')]*p12 - slotset_df.loc[i+1,('NZ')]*p12 - pPositions[0]
if slotset_df.loc[i+1,('NR')] == 1 :
neutronVi = Object3D()
neutronVi.add(Mesh(neutronGeometry, neutronMaterial))
neutronVi.position = [0,0,locN]
protonMi.add(neutronVi)
rcounter += 1
if slotset_df.loc[i+1,('NR')] == 2 :
neutronGi = Object3D()
neutronGi.add(Mesh(neutronGeometry, neutronMaterial))
neutronKi = Object3D()
neutronKi.add(Mesh(neutronGeometry, neutronMaterial))
neutronPair = Group()
neutronGi.position = [0,-n1n2/2,0]
neutronPair.add(neutronGi)
neutronKi.position = [0,n1n2/2,0]
neutronPair.add(neutronKi)
neutronPair.rotateZ(random()*2*piG)
neutronPair.position = [0,0,locN]
protonMi.add(neutronPair)
rcounter += 1
protonMi.position = [0,0,pPositions[num]]
if j == 0 : proStack1.add(protonMi)
if j == 1 : proStack2.add(protonMi)
if j == 2 : proStack3.add(protonMi)
if j == 3 : proStack4.add(protonMi)
if j == 4 : proStack5.add(protonMi)
if j == 5 : proStack6.add(protonMi)
if j == 6 : proStack7.add(protonMi)
if j == 7 : proStack8.add(protonMi)
if j == 8 : proStack9.add(protonMi)
if j == 9 : proStack10.add(protonMi)
if j == 10 : proStack11.add(protonMi)
if j == 11 : proStack12.add(protonMi)
if j == 12 : proStack13.add(protonMi)
if j == 13 : proStack14.add(protonMi)
if j == 14 : proStack15.add(protonMi)
if j == 15 : proStack16.add(protonMi)
if j == 16 : proStack17.add(protonMi)
if j == 17 : proStack18.add(protonMi)
if j == 18 : proStack19.add(protonMi)
num += 1
return
def Orient(n, stack):
# The proton stack emission plane for proStacks are oriented in the z-plane.
# Reorient the stack otherwise according to the "slotOrien" column.
if slotset_df.loc[n+1,('SlotOrien')] == "Y" :
stack.rotateX(-piG/2)
if slotset_df.loc[n+1,('SlotOrien')] == "X" :
stack.rotateX(-piG/2)
stack.rotateY(piG/2)
return
def Position(n,stack):
# Calculate the slot's position based on the the current values
# of p2p3, p12 and pE. The default values are just position markers.
loc = abs(slotset_df.loc[n+1,('P2P3')]*p2p3 + slotset_df.loc[n+1,('P12')]*p12 + slotset_df.loc[n+1,('PE')]*pE)
# Position the stack according to the presence of slotset's
# default values (make 'em one) and sign.
px = slotset_df.loc[i+1,('PX')]
py = slotset_df.loc[i+1,('PY')]
pz = slotset_df.loc[i+1,('PZ')]
if px != 0 : px = copysign(1, px)
if py != 0 : py = copysign(1, py)
if pz != 0 : pz = copysign(1, pz)
stack.position = [px*loc, py*loc, pz*loc]
return
# Assemble the atomic models.
for i in range( 19 ):
if slotset_df.loc[i+1,('Protons')] > 0 :
numPs = slotset_df.loc[i+1,('Protons')]
if i == 0 :
buildProtonStackN(numPs,i)
Orient(i,proStack1)
Position(i,proStack1) # It's (0,0,0)
grpAn.add(proStack1)
if i == 1 :
buildProtonStackN(numPs,i)
Orient(i,proStack2)
Position(i,proStack2)
grpBn.add(proStack2)
if i == 2 :
buildProtonStackN(numPs,i)
Orient(i,proStack3)
Position(i,proStack3)
grpBn.add(proStack3)
if i == 3 :
buildProtonStackN(numPs,i)
Orient(i,proStack4)
Position(i,proStack4)
grpBn.add(proStack4)
if i == 4 :
buildProtonStackN(numPs,i)
Orient(i,proStack5)
Position(i,proStack5)
grpBn.add(proStack5)
if i == 5 :
buildProtonStackN(numPs,i)
Orient(i,proStack6)
Position(i,proStack6)
grpAn.add(proStack6)
if i == 6 :
buildProtonStackN(numPs,i)
Orient(i,proStack7)
Position(i,proStack7)
grpAn.add(proStack7)
if i == 7 :
buildProtonStackN(numPs,i)
Orient(i,proStack8)
Position(i,proStack8)
grpAn.add(proStack8)
if i == 8 :
buildProtonStackN(numPs,i)
Orient(i,proStack9)
Position(i,proStack9)
grpAn.add(proStack9)
if i == 9 :
buildProtonStackN(numPs,i)
Orient(i,proStack10)
Position(i,proStack10)
grpAn.add(proStack10)
if i == 10 :
buildProtonStackN(numPs,i)
Orient(i,proStack11)
Position(i,proStack11)
grpAn.add(proStack11)
if i == 11 :
buildProtonStackN(numPs,i)
Orient(i,proStack12)
Position(i,proStack12)
grpAn.add(proStack12)
if i == 12 :
buildProtonStackN(numPs,i)
Orient(i,proStack13)
Position(i,proStack13)
grpAn.add(proStack13)
if i == 13 :
buildProtonStackN(numPs,i)
Orient(i,proStack14)
Position(i,proStack14)
grpBn.add(proStack14)
if i == 14 :
buildProtonStackN(numPs,i)
Orient(i,proStack15)
Position(i,proStack15)
grpBn.add(proStack15)
if i == 15 :
buildProtonStackN(numPs,i)
Orient(i,proStack16)
Position(i,proStack16)
grpBn.add(proStack16)
if i == 16 :
buildProtonStackN(numPs,i)
Orient(i,proStack17)
Position(i,proStack17)
grpBn.add(proStack17)
if i == 17 :
buildProtonStackN(numPs,i)
Orient(i,proStack18)
Position(i,proStack18)
grpBn.add(proStack18)
if i == 18 :
buildProtonStackN(numPs,i)
Orient(i,proStack19)
Position(i,proStack19)
grpBn.add(proStack19)
print(An) # (' 2 helium He ', 2)
print(slotlist)
#slotset_df.to_csv('out.csv')
slotset_df
The 19 slot sections of the atom building cell are now much simpler. Atom Builder can be replaced with much improved mBuilder code.
For example, Francium. TotalPs = 87, TotalNs = 85, AddedNs = 7, AlphaNs = 78, a modest 2 neutron deficit.
Francium slot 2; single neutrons are added to both ends of slot 2’s 3 alpha stack.
At the same time, there are single protons in slot two’s hook positions slot 16 and 18. The single protons align with those 2 slot 2 neutrons to form what I believe is a legitimate male-proton/female-neutron configuration. Miles has shown us the male-proton/female-proton configuration, why not then a male-proton/female-neutron? It’s similar to, but not quite the same as Miles’ diagram of Rubidium on pg 17, in “How the Elements are Built”.
Francium slot 3 has 3 added neutrons: two “left” and 1 “right”. As with slot 2, hook position slot 3 also includes a proton occupied slot 17, with its own right side neutron.
I considered converting to Miles’ Rubidium diagram at length, including my cone emission thoughts, but figured the PE/NP connections shown above make more sense. Please let me know if you (or anyone) has any objection.
Any thoughts on including an atoms’ proton and neutron slotlayout diagram? Perhaps as an addition or modification or something completely different?
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
On the subject of the SL, here's the latest SL diagram with the slot proton equatorial emission orientations: | changed to Y or X; and --- changed to Z.
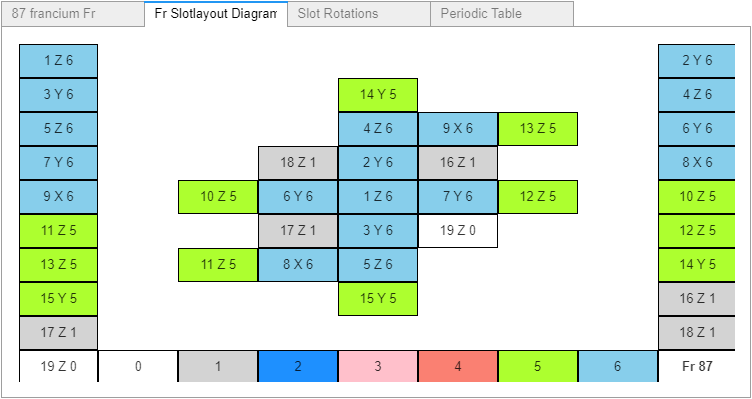
The increased information may look odd, but it's an improvement. For one thing, I think it makes visualizing the atomic +/- X, +/-Y, and +/-Z geometry clearer.
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Changes: Reviewing the code daily, I’ve made a few improvements and clean-ups.
- Code:
# Build each atom from Cr6-Elements
# atomicNumber = number # Select the desired atom from the dropdown widget or table.
grpAn = Group() # The rotation widget spins grpAn, slots 1,4 and 5.
grpBn = Group() # grpBn are the orthogonal top, bottom and hook slots.
if 1 <= number < 13:
caroSecs = 2.5
if 14 <= number < 20:
caroSecs = 4.0
if 21 <= number :
caroSecs = 6.0
proStack1 = Group()
proStack2 = Group()
proStack3 = Group()
proStack4 = Group()
proStack5 = Group()
proStack6 = Group()
proStack7 = Group()
proStack8 = Group()
proStack9 = Group()
proStack10 = Group()
proStack11 = Group()
proStack12 = Group()
proStack13 = Group()
proStack14 = Group()
proStack15 = Group()
proStack16 = Group()
proStack17 = Group()
proStack18 = Group()
proStack19 = Group()
stacks = [proStack1,proStack2,proStack3,proStack4,proStack5,
proStack6,proStack7,proStack8,proStack9,proStack10,
proStack11,proStack12,proStack13,proStack14,proStack15,
proStack16,proStack17,proStack18,proStack19]
# Dimensions # pRadius is now in In[1]
nRadius = pRadius*1.125 # Neutron radius
eRadius = 2*pRadius/10 # Electron radius
# Widget Controlled variables 1. ortho,
# 2.emR, 3. cAngleDeg, 4. pPTP, 5.p2p3Dist
pE = ortho*pRadius
emRadius = 2*emR*pRadius # 2*pE - pRadius/2
coneAngle = cAngleDeg*2*piG/360
coneHeight = emRadius*tan(coneAngle)
p12 = 2*pPTP*pRadius
p2p3 = 2*p2p3Dist*pRadius
n1n2 = (p12- 2*nRadius)/tan(coneAngle) # neutron-neutron orbital separation (y) inside the alpha
# e1e2 = -n1n2/10
p1p2 = pRadius*2.5 #0.75*p12 # pRadius*2.5
p1n1 = p12/2 #p12/2 #p12/2 # Alpha, up-down (z) proton-orbital neutron separation
p1n2 = p12/2 #pRadius + nRadius + 1.5*pRadius # In line proton and neutron
p1e1 = pRadius + 0.05*p12 # Alpha, top-bottom (z) proton-electron separation
# Proton set Geometries and Materials # Color.js Find a nice color spectrum
protonGeometry = SphereBufferGeometry(pRadius, 24, 16)
neutronGeometry = SphereBufferGeometry(nRadius, 24, 16)
electronGeometry = SphereBufferGeometry(eRadius, 8, 16)
emissionGeometryP = CircleGeometry(emRadius, 24)
emissionGeometryX = ConeGeometry( emRadius, coneHeight, 16, openEnded = True )
protonMaterial = MeshStandardMaterial(color='blue')
neutronMaterial = MeshStandardMaterial(color='red')
electronMaterial = MeshStandardMaterial(color='cyan') #'orangered','salmon','green','cyan','teal','orange'
emissionmaterial7= MeshStandardMaterial(color='gray',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial1 = MeshStandardMaterial(color='magenta',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial2 = MeshStandardMaterial(color='blue',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial3 = MeshStandardMaterial(color='pink',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial4 = MeshStandardMaterial(color='salmon',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial5 = MeshStandardMaterial(color='lime',side='DoubleSide',transparent = True,opacity = 0.20)
emissionmaterial6 = MeshStandardMaterial(color='skyblue',side='DoubleSide',transparent = True,opacity = 0.20)
if emissionType.value == 'plane':
emissionGeometry = emissionGeometryP
if emissionType.value == 'cones':
emissionGeometry = emissionGeometryX
if coloredEmissions.value == True:
emissionmaterial = emissionmaterial1
else:
emissionmaterial = emissionmaterial7
def buildProtonStackN(a,j):
# a is the total number of protons. j indicates the calling slot #.
# Each of the "a" protons in a given stack is added one at a time;
# either as the first "odd" proton added alone - plus an electron
# that is, or as the second "even" proton which is added along with
# two neutrons in order to complete an alpha: 2 p's, 2 n's and 2 e's.
num = 0
if coloredEmissions.value == True:
if a == 1 : emissionmaterial = emissionmaterial1
if a == 2 : emissionmaterial = emissionmaterial2
if a == 3 : emissionmaterial = emissionmaterial3
if a == 4 : emissionmaterial = emissionmaterial4
if a == 5 : emissionmaterial = emissionmaterial5
if a == 6 : emissionmaterial = emissionmaterial6
else:
emissionmaterial = emissionmaterial7
# Proton z positions: # The possible formula solution, rather than the list, may be in these 2 lines
# protonMi.position = [0,0,a*p12/2+(a-1)*p2p3/2-(p2p3/2+p12/2)*num] # multiAlpha solution - Not
# protonMi.position = [0,0,(a//2)*p12/2+(a//2)*p2p3/2-(p2p3/2+p12/2)*num] # pSet
if a == 1 : pPositions = [0]
if a == 2 : pPositions = [p12/2,-p12/2]
if a == 3 : pPositions = [p12/2+p2p3/2,-p12/2+p2p3/2,-p12/2-p2p3/2]
if a == 4 : pPositions = [p12+p2p3/2,p2p3/2,-p2p3/2,-p12-p2p3/2]
if a == 5 : pPositions = [p12+p2p3,p2p3,0,-p12,-p12-p2p3]
if a == 6 : pPositions = [3*p12/2+p2p3,p12/2+p2p3,p12/2,-p12/2,-p12/2-p2p3,-3*p12/2-p2p3]
def singleNeutron(neutron,position):
neutron.add(Mesh(neutronGeometry, neutronMaterial))
neutron.position = [0,0,position]
return
def pairOfNeutrons(neutron1,neutron2,position):
neutron1.add(Mesh(neutronGeometry, neutronMaterial))
neutron1.position = [0,-n1n2/2,0]
neutron2.add(Mesh(neutronGeometry, neutronMaterial))
neutron2.position = [0,n1n2/2,0]
neutronPair.add(neutron1)
neutronPair.add(neutron2)
neutronPair.rotateZ(random()*2*piG)
neutronPair.position = [0,0,position]
return
rcounter = 0
lcounter = 0
while num < a:
protonMi = Group()
protonMi.add(Mesh(protonGeometry, protonMaterial))
if emissionType.value == 'plane':
protonMi.add(Mesh(emissionGeometry, emissionmaterial))
if emissionType.value == 'cones':
emission1 = Object3D()
emission1.add(Mesh(emissionGeometry, emissionmaterial))
emission1.position = [0,0,coneHeight/2]
emission1.rotateX(-piG/2)
protonMi.add(emission1)
emission2 = Object3D()
emission2.add(Mesh(emissionGeometry, emissionmaterial))
emission2.position = [0,0,-coneHeight/2]
emission2.rotateX(piG/2)
protonMi.add(emission2)
protonMi.add(Mesh(emissionGeometryP, emissionmaterial))
electronMi = Object3D()
electronMi.add(Mesh(electronGeometry, electronMaterial))
electronMi.position = [0,-n1n2/10,-p1e1]
protonMi.add(electronMi)
protonMi.rotateZ(random()*2*piG)
if num%2 != 0:
neutronPair = Group()
neutronMi = Object3D()
neutronNi = Object3D()
pairOfNeutrons(neutronMi,neutronNi,-p1n1)
protonMi.add(neutronPair)
protonMi.rotateX(piG)
protonMi.rotateZ(random()*2*piG)
# Neutron Additions. Add neutrons where specified by the data file.
# This code adds any and all left or right neutrons during the
# addition of the "a" sized stack's first proton. pPositions[0]
# is the center for all neutrons added to that stack. Subtracting
# pPositions[0] brings the neutrons center to the center of the
# stack, so the neutron placement calculation applies equally for
# both left and right ends.
if (slotset_df.loc[i+1,('NL')] > 0) and (lcounter == 0) :
locN = ( slotset_df.loc[i+1,('N2N3')]*p2p3
+ slotset_df.loc[i+1,('N12')]*p12
+ slotset_df.loc[i+1,('NZ')]*p12
- pPositions[0])
if slotset_df.loc[i+1,('NL')] == 1 :
neutronVl = Object3D()
singleNeutron(neutronVl,locN)
protonMi.add(neutronVl)
lcounter += 1
if slotset_df.loc[i+1,('NL')] == 2 :
neutronPair = Group()
neutronGi = Object3D()
neutronKi = Object3D()
pairOfNeutrons(neutronGi,neutronKi,locN)
protonMi.add(neutronPair)
lcounter += 1
if (slotset_df.loc[i+1,('NR')] > 0) and (rcounter == 0) :
locN = ( -slotset_df.loc[i+1,('N2N3')]*p2p3
- slotset_df.loc[i+1,('N12')]*p12
- slotset_df.loc[i+1,('NZ')]*p12
- pPositions[0])
if slotset_df.loc[i+1,('NR')] == 1 :
neutronVr = Object3D()
singleNeutron(neutronVr,locN)
protonMi.add(neutronVr)
rcounter += 1
if slotset_df.loc[i+1,('NR')] == 2 :
neutronPair = Group()
neutronGi = Object3D()
neutronKi = Object3D()
pairOfNeutrons(neutronGi,neutronKi,locN)
protonMi.add(neutronPair)
rcounter += 1
protonMi.position = [0,0,pPositions[num]]
if j == 0 : proStack1.add(protonMi)
if j == 1 : proStack2.add(protonMi)
if j == 2 : proStack3.add(protonMi)
if j == 3 : proStack4.add(protonMi)
if j == 4 : proStack5.add(protonMi)
if j == 5 : proStack6.add(protonMi)
if j == 6 : proStack7.add(protonMi)
if j == 7 : proStack8.add(protonMi)
if j == 8 : proStack9.add(protonMi)
if j == 9 : proStack10.add(protonMi)
if j == 10 : proStack11.add(protonMi)
if j == 11 : proStack12.add(protonMi)
if j == 12 : proStack13.add(protonMi)
if j == 13 : proStack14.add(protonMi)
if j == 14 : proStack15.add(protonMi)
if j == 15 : proStack16.add(protonMi)
if j == 16 : proStack17.add(protonMi)
if j == 17 : proStack18.add(protonMi)
if j == 18 : proStack19.add(protonMi)
num += 1
return
def Orient(n, stack):
# proStack emission planes are initially oriented in the z-plane.
# Reorient the stack otherwise according to the "SlotOrien" column.
if slotset_df.loc[n+1,('SlotOrien')] == "Y" :
stack.rotateX(-piG/2)
if slotset_df.loc[n+1,('SlotOrien')] == "X" :
stack.rotateX(-piG/2)
stack.rotateY(piG/2)
return
def Position(n,stack):
# The default values are just used as position markers, +/- x, y or z
# with respect to (0,0,0). Use those directions but calculate the slot's
# position magnitude based on the values of p2p3, p12 and pE.
loc = (abs(slotset_df.loc[n+1,('P2P3')]*p2p3
+ slotset_df.loc[n+1,('P12')]*p12
+ slotset_df.loc[n+1,('PE')]*pE))
# Determine the direction of the stack according to the presence and sign
# (+/-) of slotset_df's occupied (x,y,z) positions. Either a value or a
# zero, '0'. If there's a value, let it be given the position magnitude.
px = slotset_df.loc[i+1,('PX')]
py = slotset_df.loc[i+1,('PY')]
pz = slotset_df.loc[i+1,('PZ')]
if px != 0 : px = copysign(1, px)
if py != 0 : py = copysign(1, py)
if pz != 0 : pz = copysign(1, pz)
stack.position = [px*loc, py*loc, pz*loc]
return
# Assemble the atomic models.
#grpAn: Carousal 1,6,7,8,9,10,11,12,13
anSlots=[0,5,6,7,8,9,10,11,12]
for i in range( 19 ):
if slotset_df.loc[i+1,('Protons')] > 0 :
numPs = slotset_df.loc[i+1,('Protons')]
buildProtonStackN(numPs,i)
Orient(i,stacks[i])
Position(i,stacks[i])
if i in anSlots:
grpAn.add(stacks[i])
else:
grpBn.add(stacks[i])
print(An) # (' 2 helium He ', 2)
print(slotlist)
#slotset_df.to_csv('out.csv')
slotset_df
1. After having added almost 400 new neutrons, there were three instances where either individual neutrons or neutron pairs were being created - in both the left and right ends of a stack as well as in each alpha inside each stack. Added two new functions inside buildProStackN() shown above, singleNeutron() and pairOfNeutrons() which replace those coded additions with code saving function calls.
2. Created “stacks=[]”, a list made up of the existing proStacks1 through proStack19 'Groups'. I was then able to iterate through stacks to reduce the "Assemble the atomic models" section from 19 if statements down to just one, the last one in the code above.
Reviewing to see if I can apply a similar ‘iterable’ approach to any of mBuilder’s many other lists.
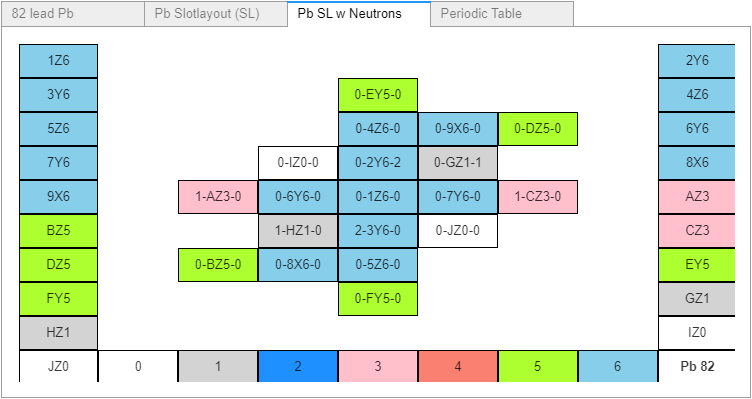
3. As you may have expected from my previous post, am attempting to come up with a modification to the existing SL diagram to include neutrons, or keep the existing SL as is and give this SL diagram its own tab. In any case, the button labels must be as compact as possible.
In the center and side columns we still have the same core three values: a. ‘SlotNumber’, b. ‘SlotOrien’ and c. ‘Protons’, the color-coded number of protons in that slot. The three values are displayed together without spaces. I’ve ensured there can only be three characters by replacing the one or two digit 19 slot decimal numbers with a single alpha-numeric, counted 1-9 followed by A-J. 1-J, 19 unique characters.
The buttons in the SL’s center atomic configuration also include each stack’s left and right end neutrons. Note that Pb slot 2 contains 2 right side neutrons, 0-2Y6-2. Some browser view size/settings will cut off the ends of overly long button descriptions. Knowing the neutron SL character code, I would further delete the 2 dashes to give each button just 5 A/N characters, Pb slot 2 would then show 02Y62. I don’t see any ambiguity with such a 5 digit label, leaving the side columns at three characters pretty much shows how and where the neutron are added.

Another thing. Now that there’s a complete set of organized data, would you approve my attempting to program the Cr6-Elements.csv’s SL cells? I’ve created plenty of excel computations and diagrams, and have used excel to solve math problems as you might use R. Automating your excel SL diagrams (within each 19 row atom) with colors, numbers and strings is something new but it can’t be as difficult as pandas.
Ready to switch Atom Builder with mBuilder and starting a new multi-atom builder, not at all sure how switching versions work.
How about di-atoms for starters?
.
LongtimeAirman- Admin
- Posts : 2078
Join date : 2014-08-10
Chromium6 likes this post
Page 3 of 13 •  1, 2, 3, 4 ... 11, 12, 13
1, 2, 3, 4 ... 11, 12, 13 
 Similar topics
Similar topics» Cr6's Periodic Table
» RSC's Periodic Table for Comparison
» Periodic Table of Toxic Elements
» Happy sesquicentennial to the periodic table of the elements