Miles Periodic Table with Standard Periodic Table reference
Page 4 of 12 •  1, 2, 3, 4, 5 ... 10, 11, 12
1, 2, 3, 4, 5 ... 10, 11, 12 
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Sorry for a lack of input or feedback on my part. Just been busy.
Really like the added notation for slots.
Di-atoms sounds good to you are leading us all on this build of atom bonding configurations.
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Glad you’re alive Cr6. You didn’t mention automating Cr6-Elements.csv? Well, that’s Ok, I may need to reconsider my offer. Ultimately, it needs its own working slotlayout, but I no longer have excel. On my latest machine I’ve been using Libre Office Calc to make all the csv file changes. It may take a bit longer learning Calc. I’ll put it on a back burner for the time being.
Changes:
1. Switched from the previous perspective camera to a “combined” camera, and added the controls allowing a user to switch between either orthographic or perspective views. Above is an orthographic view of radium, in which all protons or all neutrons appear the same size, with no foreshortening which is present in perspective mode. Both views are obtained using the pythreejs orbital controls. It teased me by waiting till I was almost done coding the camera change before the non JSON compliant User Warning reappeared.
2. I went ahead and replaced AtomBuilder (AB) with a copy of mBuilder, and named it AtomBuilder2. AB has been moved to the notebook 2 folder alongside mBuilder. mBuilder carries on as the day-to-day working program intended to develop a 3D charge field model rendering of molecular matter selected by the user.
Since we're at the top of page 7, here's the github location where the latest Jupyter notebook Atom Builder project can be found. https://github.com/LtAirman/ChargeFieldTopics

.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
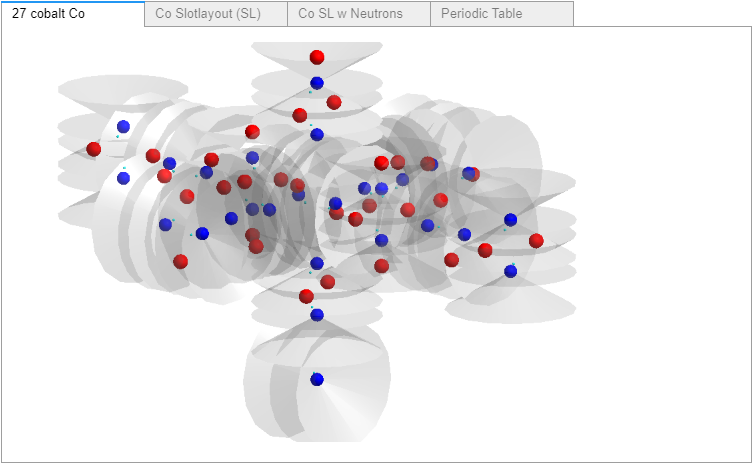
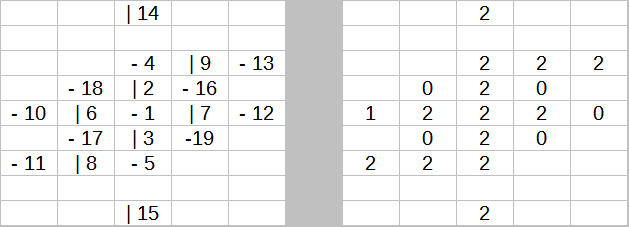
Change 1. A before and after of Cr6-Elements.csv SL for Cobalt (27). All 90 SLs were changed from containing the same slot orientations and slot numbers on the left, to each atom’s unique slot proton counts (0-6) on the right. The simplest SL yet, going from two to just one of the 3 central SL coded values used in AtomBuilder2 (AB2). Note the lack of any “color coding”, that’s because csv files do not support color coding.
That’s not what I’d intended, I thought I’d be able to automate Cr6-Elements.csv and include all three bits of information using some combination of formulas, text strings, color coding and conditional formatting. On the “back burner”, I began by updating my copy of Libre Office, then downloading the Getting Started Guide and the 7.2 Calc Guide. Did some reading and worked some examples, over and over again, developing libre calc conditional coding muscle memory if nothing else. Over and over, I was unable to save any formula or formatting changes when reloading the file, except when I ran the relative reference formulas identifying where to find the number of protons in each of the SL’s 19 slots first (in the slot numbered row and “Protons” column). Then, after restarting the file, the formulas again disappeared but the proton count output changes remained. Positive changes are always welcome.
Coding is routinely humbling. I’ll usually try some alternative that I’ve seen or had occured to me, then find it doesn’t work anywhere near how or when I thought it should. I must be extremely careful and organized when attempting small logical changes while regularly repeating to myself, the code isn’t screwing-up, I am. This time it was a slow realization that Cr6-Elements.csv cannot be automated, it contains no formulas, color coding or formating, csv files do not support such things. They would instead be found in, say, an excel Cr6-Elements.xlsx file. Using pandas and dataframes, when said Cr6-Elements.xlsx is imported as a csv - coma separated values file, all the higher functionality and formatting of the xlsx file is not included in the csv’s text output.
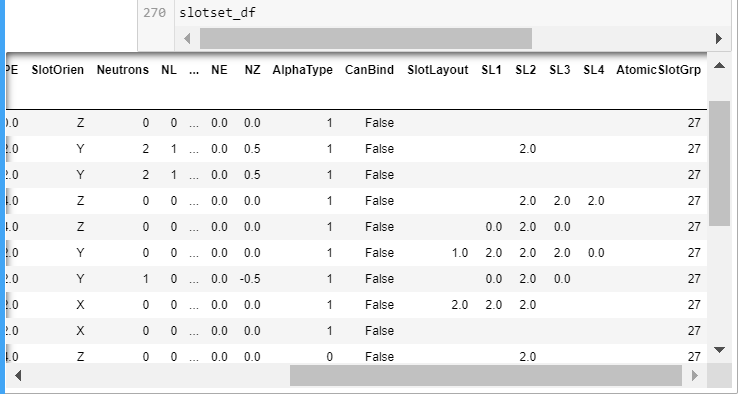
Change 2. The SL cells with multiple data (SL numbers and pipes or dashes) were gone so I didn’t need to worry about them as I did in the past. AB2 and mBuilder now read and display Cr6-Elements.csv’s 90 SL diagrams in a sort of recursive touch. The original empty spaces above and below the SLs were read and displayed as a multitude of NaNs by pandas, I converted back to spaces with df = df.fillna(" "), giving the SL a nice viewport in the slotset_df printed output after the atom building cell.
Those changes have satisfied any need I may have had for any data file “automation”. I’m out of any additional improvement ideas, and declare AB2 well started. Am attempting to turn my thoughts to coding changes that may be necessary for multi-atoms.
Di-atoms sounds good to you are leading us all on this build of atom bonding configurations.
I beg to differ. Believe it or not, all my best work is for others – not for myself. I’m all about mission, service, teamwork, commitment and accountability. Left to myself I’d take a break then find another mission. With respect to multi-atoms, may I remind you, you’re still the boss. This project would have gone nowhere without your work, ideas, and “leadership”. You’ll recall Nevyn built another program which displayed all kinds of compound atomic structures well beyond AtomicViewer’s output. Well beyond anything I’m capable of. I certainly didn’t use it or study his structures at any length, but I believe you did. I’m learning but you’ve always had a much better understanding of Miles’ atomic models than I do. How to organize it? What other table columns should come into play? And where does machine learning come in? Sorry I'm such a nag.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
However, we could serialize each key column in pandas for allowing joins with other columns. If we need to add in rotation or charge field directions. The dataset could get really large. This is the difference between what do you want to see in the gui with allowed dynamic calcs verses what is allowed with data set value lookups on the dataset.
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Thanks Cr6. If you have any algorithm ideas you want me to try, just sing out.Cr6 wrote. Awhile back I was looking at using common two-three molecule formations (e.g. SO2) with the Slot formations to see if bonds could be allowed with Miles' Periodic table using allowed-not allowed structures with Miles' atoms. … I was looking at building an algo just to say these stick against the wall and these don't with the slot layouts
I was quickly overwhelmed by sheer numbers alone. What? Tens of thousands(?!) with more molecules discovered every day? On the c.f. side, the variable atomic/molecular configurations possible under different charge field conditions, i.e. each hydrogen’s different proton position and neutron orientation within a single water molecule * depending on whether from whether the main charge flow enters either the top or bottom here on Earth, I find even more daunting. Everything is dynamic. Attempting to create some common two-three molecule formations from a given list – i.e. fixed, non-dynamic models is probably the only way I can proceed for now.
For the record, this old below average chemistry student is convinced that the charge field – photon charge flow channeled and recycled through particles, atoms and molecules, is the single theory that explains all matter behavior. I’ve happily rejected the premise that everything can be explained with equal and opposite proton and electron charges. Given the acknowledged 1840X mass difference between them alone, electron cloud or not, I see the existing mainstream bonding theories as absurd. Physical chemistry needs to be tossed out and completely transformed. Nevyn and Miles have expressed the same sentiment. Like it or not, this project forces me to learn more on the subject. I’ll need plenty more understanding and an ongoing review of Miles’ SECTION 9: THE NUCLEUS papers. Feel free to point out my errors.
Googling a good warm up question, “What are the most common elements in the universe?”, returned plenty of hits, including:
Science Notes, What is the most abundant element in the universe, 27 October 2020 (updated 10 May 2021),
https://sciencenotes.org/what-is-the-most-abundant-element-in-the-universe/
Anne Helmenstine provided a chart with the answer: Hydrogen: 73.9%, Helium:24.0, Oxygen: 1.0%, Carbon:, 0.5%, Neon, 0.1%, Iron: 0.1%, Nitrogen: 0.1%, All Others: 0.3%.
Anne Helmenstine wrote. Hydrogen is the most abundant element in the universe, followed by helium. Oxygen accounts for about 1 percent. All the other elements together only account for a bit more than one percent!”.
Below is a chart that puts those numbers into better perspective. Currently explained by a slow conversion of hydrogen into helium in the cores and across stellar generations. It also happens to match our atomic range in AB 1 or 2 very well. Uranium can be a special case. In addition to wiki, I think I’ve seen it with different text elsewhere before.
https://www.forbes.com/sites/startswithabang/2020/05/25/this-is-where-the-10-most-common-elements-in-the-universe-come-from/?sh=519c9966d24b
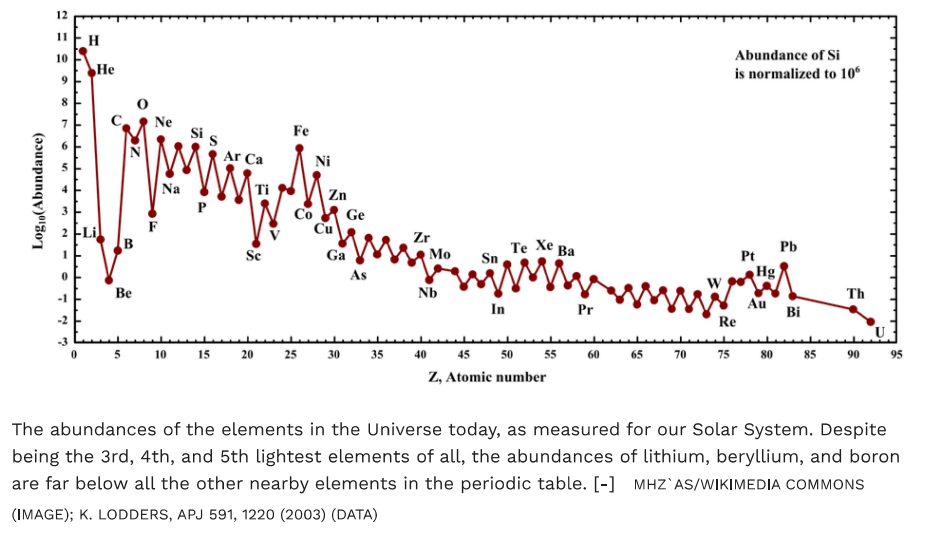
I believe these are total atomic mass percentages, and not total numbers of atoms. One conceptual question might be – can the charge field explain those up/down zig-zaging abundances? Another, why do Li, Be and B constitute an unusual single low abundant zag compared with the main trend of the elements? What is it about iron (F3, 26) that makes it especially abundant? Where’s promethium (Pm, 61). Examining our atomic models against this list might make a perfectly good project using AB2.
Ok, molecules, the main question next - “What are the most common atomic compounds in the universe?” Many less hits this time, google returned mostly the same results as my previous question. Special thanks to Uncle Al for his reply with two good links, A and B included below, from a chemistry stack exchange discussion.
Leading with some spectroscopy background.
https://en.wikipedia.org/wiki/List_of_interstellar_and_circumstellar_molecules
Wikipedia
Airman. I wish I could point to all the non-charge field errors contained in that paragraph. Of course nobody could do it as well as Miles. “when one of the molecule's electrons moves between molecular orbitals” is wrong, I don’t believe the molecule’s ambient quantization levels being observed necessarily involve any electron re-distribution, electrons are just along for the ride, carried by the charge streams, unable to penetrate neutrons or protons. The rest of it looks fairly good to me. Notable in that it admits absorption or emission of photons at various energy levels. Not single photons, but an average quantization of all the photons recycling through all that molecule’s various nuclear component particles as well. Glad to see Spectroscopy can also provide spin information.Background[edit]
The molecules listed below were detected through astronomical spectroscopy. Their spectral features arise because molecules either absorb or emit a photon of light when they transition between two molecular energy levels. The energy (and thus the wavelength) of the photon matches the energy difference between the levels involved. Molecular electronic transitions occur when one of the molecule's electrons moves between molecular orbitals, producing a spectral line in the ultraviolet, optical or near-infrared parts of the electromagnetic spectrum. Alternatively, a vibrational transition transfers quanta of energy to (or from) vibrations of molecular bonds, producing signatures in the mid- or far-infrared. Gas-phase molecules also have quantised rotational levels, leading to transitions at microwave or radio wavelengths.[1]
A. Observed Interstellar Molecular Microwave Transitions
Recommended Rest Frequencies (nist.gov)
https://physics.nist.gov/cgi-bin/micro/table5/start.pl
NIST Recommended Rest Frequencies for Observed Interstellar Molecular Microwave Transitions
by Frank J. Lovas
Airman. And the explanatory link.
Observed Interstellar Molecular Microwave Transitions
https://www.nist.gov/pml/observed-interstellar-molecular-microwave-transitions
NIST Standard Reference Database 116
Last Update to Data Content: September 2009
Airman. From which I’ve gleaned two lists: 273 molecule formulas, and 118 “empirical” intestellar molecules. Where are the frequencies? The only one I see is 20MHz.
B. The Cosmic Ice Laboratory - Interstellar Molecules
Containing 12 Tables identifying “Molecules with (2 through 13) atoms,
https://science.gsfc.nasa.gov/691/cosmicice/interstellar.html
Interstellar Molecules - The Cosmic Ice Laboratory - Sciences and Exploration Directorate - NASA's Goddard Space Flight Center
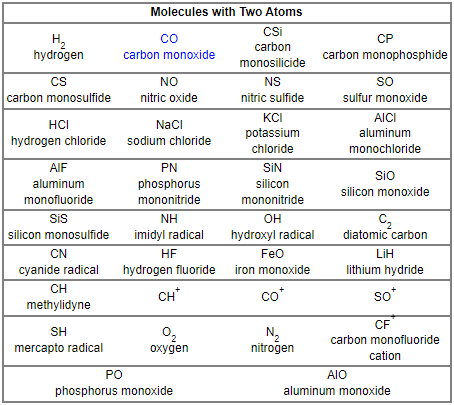
Airman. From which I’ll share the first two of twelve tables identifying Molecules with Two – Thirteen Atoms.
Airman. Unless you have any other suggestion or objection, I think I’ll try working with these two charts.
Cr6 wrote. It is always easier to add new columns for category logic than to parse a unique column for values.
Airman. Hallelujah brother. Quantized transition frequency columns might be nice.
Cr6 wrote. However, we could serialize each key column in pandas for allowing joins with other columns. If we need to add in rotation or charge field directions. The dataset could get really large. This is the difference between what do you want to see in the gui with allowed dynamic calcs verses what is allowed with data set value lookups on the dataset.
Airman. In order for the spin animations to work I’ll probably need to add spin directions before any joining occurs.
I see joining according to serialized key rows (slots), but I don’t see joining w.r.t. serialized key columns yet. I suppose key columns might identify such things as phase transitions between energy ranges?
jupyter-widgets/pythreejs. In the issues section and message traffic at gitHub’s pythreejs/jupyter bridge, a couple of individuals have indicated their desire to upgrade pythreejs to threejs version r112. vidartf has replied, welcoming them, glad for and willing to help, indicating his own intent to update pythreejs to work with ipywidgets 8. I’ll report any other significant activity as/if it happens.
Possible action items. I should have this mentioned months ago; of course you noticed that the beryllium (Be, 4) I modeled as a single slot alpha and 1 each orthoigonal single protons in slots 2 and 3 ( -- | : | -- ) is not even close to Miles’ Be diagram, ( | . | : | : | ) - 4 parallel protons with one or two neutrons between adjacent protons. I didn’t follow that image/example because I couldn’t figure out how to get it into our “allowed” 1-19 slot model format. I don’t recall any other atoms with as drastic a image/model difference. Furthermore, the possibility of neutrons between adjacent parallel alphas or protons greatly expands the number of neutrons that can be found in a given atom. Do you think we need any action items to identify required AB2 corrections?
*
324. The Hydrogen Bond. http://milesmathis.com/water2.pdf Including a diagram of water. 10pp.
340. The Fourth Phase of Water, part 1. http://milesmathis.com/poll.pdf I look at Gerald Pollack's book, analyzing polywater, the exclusion zone, and charge channeling. 15pp.
341. THe Fourth Phase of Water, part 2. http://milesmathis.com/poll2.pdf I show how Pollack's theory fails, and offer a replacement. 15pp.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
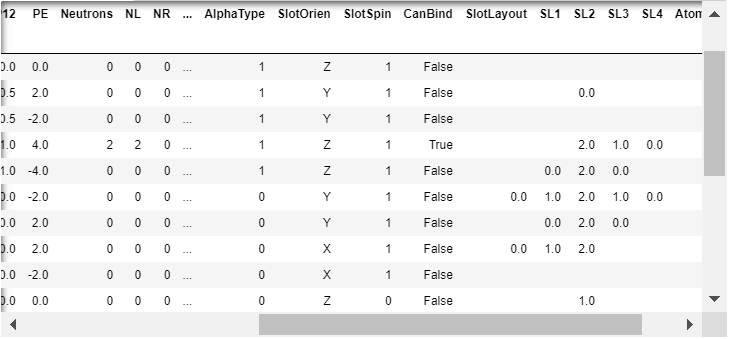
The ‘SlotSpin’ column has been added.
Staus update. Truth be told, I don’t know how I’ll proceed. The paperwork (status updates) is a constant that must show progress, so do something. The updates help organize my thoughts and things do seem to get done. As long as everything else continues to work properly, most any change can be undone. Let the first atom be A, and the second atom be B. In preparation for adding B to the rendered scene, here are the latest changes:
1. As you’ve reminded me, finally added a ‘SlotSpin’ column to Cr6-Elements.csv. To the right side of ‘SlotOrien’. Both columns are moved to the right side of ‘AlphaType’. All occupied slots can have either a positive or negative 1 value. All values are initially set to 1 and have yet to be accessed by the ‘atom building’ cell. Not at all unusual, I needed to change the code again (reflected in the github repository commit history) after finding a brand new error; this time a blank column - now removed.
- Code:
rowstart = 19 * (atomicNumber - 1)
#rowstop = rowstart + 19 # Used with the iloc slicing
#slotset_df = df.iloc[rowstart:rowstop, 8:33].set_index('SlotNumber')
rowstop = rowstart + 18 # Used with the loc slicing
slotset_df = df.loc[rowstart:rowstop, 'SlotNumber':'AtomicSlotGrp'].set_index('SlotNumber')
2. Both AB2 and mBuilder access the csv file and needed a corresponding column index number change. Normally I wouldn’t mention it, but for the second time I made the notable improvement of converting that difficult and troublesome (thwarted prior attempts) single numerical iloc[] index into an easier to read and maintain column labeled loc[] index.
3. Added an initial molecules dictionary {'H2':1,'C2':2,'N2':3,'O2':4}.
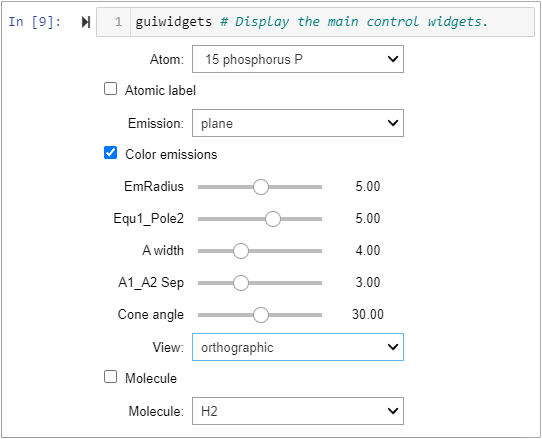
The current ipywidget controls graphic user interface (gui).
4. Added 2 control widgets: ‘Molecule’ checkbox, and ‘selectMolecule’ dropdown widgets.
5. Added a temporary print('moleculeOrNot…) output, a true or false status indicator for coding convenience.
6. Renamed the single atom’s 2 spin groups: ‘grpAn’ to ‘grpA1’, and ‘grpBn’ to ‘grpA2’. Made the corresponding variable name changes to the ‘rendered scene’ and ‘rotation widgets’ cells.
7. Added the 2nd atom’s spin groups: grpB1 and grpB2. Made those group additions to the ‘rendered scene’ cell.
8. Renamed A’s 19 proton stack groups ‘stacks[] = [proStack1, …, proStack19]’ to ‘stacksA[] = [proStackA1, …, proStackA19 ]’.
9. Added the second atom’s list of proton stack groups slots, ‘stacksB[] = [proStackB1, …, proStackB19 ].
10. Renamed Atom 1 spin animation variable (_track, _clip, and _action) prefixes to spinA1 thru spinA19.
11. Added ‘Atom 2 spin animation’ cell, with widget control variable (_track, _clip, and _action) prefixes spinB1 thru spinB19.
Better start my next update as soon as I can.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Status update. Not much tangible progress to report this week.
molecules = {'H2':1,'C2':2,'N2':3,'O2':4}
moleConfig = [(2,1,1), (2,6,6), (2,7,7), (2,8,Cool]
1. Renamed the ‘Elements’ cell to ‘Elements and Molecules’. Moved the molecules dictionary from the 'gui widgets' cell to the ‘Elements …’ cell, and added moleConfig, a list of 4 lists each with 3 integer values: 1. The total number of atoms in the molecule; 2. The atomic number for the first atom; and 3. The atomic number of the second atom. The dictionary and list of lists are used in the 'gui widgets' and 'Atom data' cells.
2. Before adding a second atom's: 'slotsetB', and dataframe 'slotsetB-df; renamed all the first atom occurances of: 'slotlist' to 'slotlistA'; and 'slotset_df' to 'slotsetA_df".
- Code:
# Atom data.
# ... The Operating instructions are omitted.
firstAtom = False
secondAtom = False
thirdAtom = False
moleAtoms = [firstAtom, secondAtom, thirdAtom]
# Atom data. After making an element or molecule change,
# obtain the selected set of atomic data.
# Selected Atom or molecule
if moleculeOrNot.value != True:
# Must be a single atom, so use the AB2 atom data central code.
An = elements[number-1]
atomicLabel = An[0]
atomicNumber = number
elementlist = df['Element'].drop_duplicates().array
atom = elementlist[number-1]
atomsymbol = dlist[number-1]
rowstart = 19 * (atomicNumber - 1)
rowstop = rowstart + 18 # Used with the loc slicing
slotsetA_df = df.loc[rowstart:rowstop, 'SlotNumber':'AtomicSlotGrp'].set_index('SlotNumber')
slotlistA = list(slotsetA_df.loc[:, 'Protons'])
"""
print('number = ',number)
print('An = ',An)
print('atom = ',atom)
print('atomicNumber = ',number)
print('atomsymbol = ', atomsymbol)
print('atomicLabel = ',An[0])
"""
#print('slotlistA = ',slotlistA)
#slotsetA_df
#slotsetA_df.info()
else :
# A molecule. Diatoms for the time being.
# Beginning with the number of specific atoms. numberM
# identifies the molecular number (1-4) from this set.
#molecules = {'H2':1,'C2':2,'N2':3,'O2':4}
#moleConfig = [(2,1,1), (2,6,6), (2,7,7), (2,8,8)]
numberOfAtoms = moleConfig[numberM-1][0]
for i in range(numberOfAtoms):
moleAtoms[i] = True # first, second, and maybe third atom.
if i == 0:
# Atom 1 of 2.
number1 = moleConfig[numberM-1][1]
An1 = elements[number1-1]
atom1 = elementlist[number1-1]
atomicNumber1 = number1
atomicLabel1 = An1[0]
atom1symbol = dlist[number1-1]
rowstart1 = 19 * (number1 - 1)
rowstop1 = rowstart1 + 18 # Used with the loc slicing
slotsetA_df = df.loc[rowstart1:rowstop1, 'SlotNumber':'AtomicSlotGrp'].set_index('SlotNumber')
slotlistA = list(slotsetA_df.loc[:, 'Protons'])
#print(slotlistA)
#print(slotsetA_df)
#slotsetA_df.info()
if i == 1:
# Atom 1 of 2.
number2 = moleConfig[numberM-1][2]
An2 = elements[number2-1]
atom2 = elementlist[number2-1]
atomicNumber2 = number2
atomicLabel2 = An2[0]
atom2symbol = dlist[number2-1]
rowstart2 = 19 * (number2 - 1)
rowstop2 = rowstart2 + 18 # Used with the loc slicing
slotsetB_df = df.loc[rowstart2:rowstop2, 'SlotNumber':'AtomicSlotGrp'].set_index('SlotNumber')
slotlistB = list(slotsetB_df.loc[:, 'Protons'])
#print(slotlistB)
#print(slotsetB_df)
#slotsetB_df.info()
print('numberM = ', numberM, ', numberOfAtoms = ',numberOfAtoms)
print('moleConfig[numberM-1] = ',moleConfig[numberM-1] )
print('moleAtoms = ',moleAtoms)
print('number1 = ', number1,', number2 = ',number2)
print('An1 = ', An1,', An2 = ', An2)
print('atom1 = ', atom1, '. atom2 = ',atom2)
print('atomicNumber1 = ',atomicNumber1,'atomicNumber2 = ', atomicNumber2)
print('atomicLabel1 = ', atomicLabel1,', atomicLabel2 = ', atomicLabel2)
print('atom1symbol = ',atom1symbol,', atom2symbol = ', atom2symbol)
print('slotlistA = ',slotlistA)
print('slotlistB = ',slotlistB)
3. Expanded the 'Atom data' cell (included above) beyond a single atom to include the selected molecule's atomic data. The desired data is provided, but the code needs better organization. Many python methods are still new to me. Developing the ‘Atom data’ cell makes it clear I should continue reviewing and improve my understanding of python functions, lists and dictionaries.
4. One intangible included reviewing those sections (python functions, lists and dictionaries) in a free electronic book I hadn’t seen before – Automate the Boring Stuff. https://automatetheboringstuff.com/
5. Another not exactly intangible, while reviewing H2, (the first molecule I’ll attempt to display), I identified an error with my charge field alpha model.
In Diatomic Hydrogen*, Miles explains that of the eight possible configurations of two electrons and two protons forming parahydrogen and orthohydrogen, the configurations with two electrons between the two protons cannot occur since the electrons’ charge vortices interfere with the charge channel between the protons and so prevent a charge bond.
From Deuterium and Tritium**, with respect to He, Miles wrote.
Embarrassment and dismay, I incorrectly placed the two electrons inside the alpha. I suppose I should pull at least one of the electrons out and place it in orbit about the alpha’s charge intakes.We need to know how these 2H's actually fit together, neutrons and all. Do they fit end-to-end, as drawn above, or do they go side-by-side, as I have drawn them before? Due to the known compactness of the He nucleus, as well as to the compactness of all other nuclei, it must be the latter. In previous calculations on larger nuclei which are composed of He nuclei, I have found the height and width of the nuclei by assuming my He sandwich, with the neutrons side-by-side. This gave me the correct ratios to match current data, so I assume that as strong indication of my first diagram of the alpha.
On the other hand, the two neutrons present are not spinning in direct line with the protons as in Diatomic Hydrogen, but orbiting around the direct proton to proton charge channel thereby creating a pair of two rotating charge vortices in both directions. I imagine two electrons can find perfectly stable positions within an alpha by orbiting each neutron’s charge intake. Your consideration in this matter would be welcome. And please, feel free to point out my errors.
*
332. Diatomic Hydrogen. http://milesmathis.com/diatom.pdf My new charge bonding explains this much better than electron sharing. Plus an analysis of spin isomers. 9pp.
**
333. Deuterium and Tritium. http://milesmathis.com/deut.pdf I show the nuclear diagrams for these, as well as for heavy water. 20pp.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

An alpha, Helium, He. More specifically, He4 is the isotope with 2 protons and 2 neutrons. Now with exterior electrons orbiting the top and bottom alpha charge intakes. True for all alphas in all atoms.
Another mostly intangible week. Only 2 code changes to the 'Build the Atom' cell in both AB2 and mB:
1. Created a new emissionMaterialList and changed the 'emissionmaterial' assignments to refer to that list.
emissionMaterialList = [emissionmaterial1,emissionmaterial2,emissionmaterial3,
emissionmaterial4,emissionmaterial5,emissionmaterial6,emissionmaterial7]
Emissionmaterial1-6 are the six different colors coded for stacks of 1-6 protons. Emissionmaterial7 is the grey colored emission field alternative shown above.
2. Also shown above, moved the two interior electrons from any and all alphas to the two exterior alpha charge intakes. Positioned with respect to the closest proton, just a sign change for the electron’s z value, probably the smallest change in the project’s repository.
In the intangible category:
A. Reviewed another “functions” chapter in another free python book by Al Sweigart, Beyond the Basic Stuff With Python, Best Practices for Writing Clean Code.
Chapter 10 - Writing Effective Functions (inventwithpython.com)
https://inventwithpython.com/beyond/chapter10.html#
No great insights. His molecule *args and **kwargs example isn’t particularly applicable, but he does offer plenty of suggestions for improvements. Plus chapters on Git and Object Oriented programming. I went ahead and purchased a paper copy from a bookstore. I much prefer reading - from paper, like before going to sleep. By the way, copies of Miles’ books: The Un-Unified Field, The Incorporation of Light and Navigating the Nucleus have been on my nightstand since 2010, 2011 and 2013. Time sure flies.
B. Continued reviewing Deuterium and Tritium, or 2H and 3H in accordance with the hydrogen isotope labels with one and two neutrons. A great paper, foundational to the charge field atomic models and this project. Gives me plenty to consider.
I’ve generally understood the alpha to be the basic charge field atomic unit, which Miles has referred to as the first true element – a stable collection of atomic particles. Hydrogen, H, the isotope protium, is just a proton without a neutron. 2H, A proton with a neutron is much more interesting. 2H’s proton and neutron (and electron) create a charge channel stronger than a proton alone, that can attract a baryon – either a proton or a neutron, or even another 2H. A small amount of 2H is found in water on Earth and elsewhere, and it is commonly involved in atomic reactions. Miles explains 2H is not necessarily formed inside suns, and goes on to explain how oxygen17 in water molecules can naturally create a pair of 2H’s. He4, the alpha itself is a bonded pair of two opposing 2H’s. With particles: proton and anti-proton, neutron and anti-neutron, electron and positron. 3H is a proton with a neutron at each proton pole. 3H is unstable. When the neutrons align, the combined charge current through the proton adds to 1.37. Well over a third more charge current than a single proton naturally channels. Especially in an unbalanced charge field, one or both of 3H’s neutrons will be kicked out.
Please pardon the review, I’m seeing stuff I don’t recall seeing before.
Displaying molecules should require more spin accuracy. Any slot joining two atoms should be able to display two opposing or complementary spin directions at the same time. At present, as you know, this project isn’t based on the alpha. Its written such that the basic atomic unit is any given slot, one of up to 19 comprising the atom’s slotlayout (SL) configuration. The occupied slot’s contents vary: 0 to 6 protons, neutrons and electrons. Each and every particle in that slot spins at the sane rate as a single object about that slot’s proton stack charge channel axis. The only way to discern spin is to observe offset neutron and electron rotations about the main charge channel. As such, with 19 spin controls the code provides an illusion of an atom’s actual spin motions.
Currently wondering whether or how to go about dividing each slot into independent subgroups such as 2H? I suppose the spin animations will become completely unwieldy.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

mBuilder now displays contra spins, positive and negative for each of the up-to 6 possible particle groups within any given slot. Impossible to confirm with a single image, even worse if neutrons aren’t included. With the rotations active, each proton still has an +/- axial spin as well as an electron, fixed to that +/- axial spin, “orbiting” the proton’s charge intake.
All prior atomic spin animations were positive. The recent spin additions intended for a second atom have been repurposed to provide a complete set of negative spins. Within a slot, each proton’s spin direction should be chosen as either positive or negative in accordance with the alpha configuration of that stack – working on it, more below. Neutrons sharing an orbit within an alpha cannot be allowed to rotate thru each other in opposite directions, so alpha neutrons are added as a diametrically separate pair, both with the same spin direction, fixed to the alpha’s second proton center and axial spin, along with the alpha’s second electron.
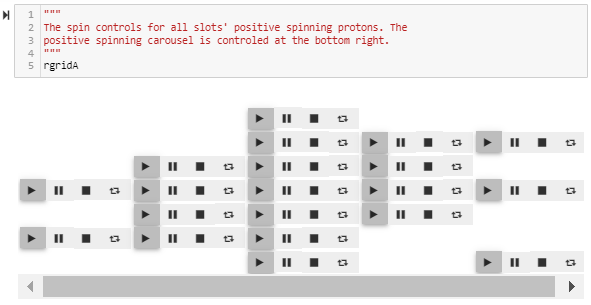
The positive spin controls are shown just before the displayed tab, and the negative spin controls are shown just after. Turning on a slot’s positive spin will animate all the particles within that slot with a positive spin direction. Same for the negative spinners. With two switches, all the particle groups are animated. The carousel is now turned on or off by a new spin controller at the bottom right corner of the positive spin controls.
Continued in the next post.
.
Last edited by LongtimeAirman on Fri May 27, 2022 5:06 pm; edited 1 time in total
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Continued from the previous post. Four images forced me to break it into two parts.
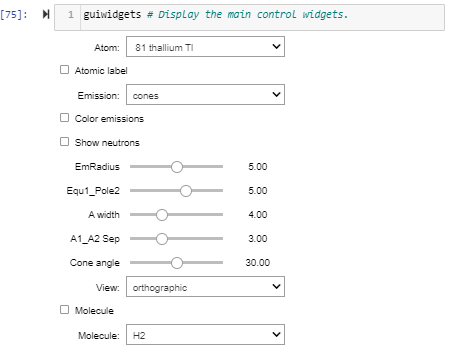
The atomic motions demonstrated by just two opposing proton axial spins are still an over-simplified illusion, but the illusion is much improved. One could easily see each slot’s ‘ganged’ single rotation. With the addition of contra-spins the animation of Thallium above looks like a stirred-up bee hive. All those spins look good, otherwise all that motion is very distracting. Added a new gui checkbox, ‘Show neutrons’ to let the user display an atom with or without them.
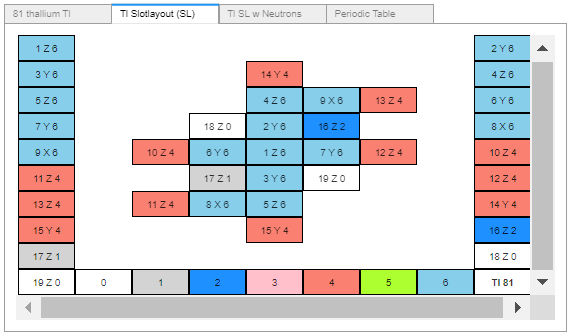
The second, negative spin set has been added to mBuilder but not AB2. Before doing so, the Cr6-Elements dataframe needs changing and I’m not sure how. It currently includes a single ‘SlotSpin’ column, with a possible +/- 1 value that applied to the entire slot. That data needs to be expanded to include the slot’s up-to 6, particle groups, particle order and spin direction.
Ordering my thoughts with the intent of encoding the 1. spin direction and 2. Up-to 6, Single slot particle groups.
Spin direction. My current understanding - with the usual caveats and likely mistakes follows. For single spin directional slots, no coding rules were necessary, all slots spun in the same direction. I was ready to rotate the single spin 180 degrees to make a second spin direction. That would have fit the data frame’s single ‘SlotSpin’ column containing a +/- 1 value. The problem is, as I understand it, given a predominate charge field direction – say up, a single spin is akin to selecting only a right threaded screw pointing up, ok, you can also point the screw down. Two spins is like having both right and left threaded screws - that can be pointed up or down. As with diatomic hydrogen, there are two spin alternatives (left or right) and two charge channel directions (up or down), indicated by the presence of an electron at the proton’s primary charge intake. Four possible spin direction/charge direction alternatives for a single proton/electron. With two aligned proton/electron pairs there are eight possible combinations, only four of which are allowed. i.e. All combinations with two interior electrons are disallowed.
Up-to 6, Single Slot particle groups:
1. proton.
2. proton + electron.
3. proton + electron + neutron.
4. neutron.
5. 2 neutrons
6. 2 protons + 1 neutron.
7. 2 protons + 2 neutrons + 2 electrons, (single alpha).
Enough for now.
Completely unwieldy? All spin animations require clicking on each and every “on” switch, 20 positive and 19 negative. Ideally, each particle: proton, neutron and electron, should have its own axial spin and set of 39 switches, groups, lists, etc. – I think I’ll pass for the time being. At minimum, with two proton spins, each additional atom will add the same possible 39 spin groups. Not completely unwieldy, but close. With up to 19 occupied slots, each with vaying particles and spins, *args and **kwargs should come in handy. Till then I’ll keep trying to to gang the spins switches together somehow. I couldn’t find the readTheDocs spin animation example I used as a model for this project yesterday. Things change so quickly. https://ipywidgets.readthedocs.io/en/stable/examples/Widget%20List.html
GitHub jupyter-widgets/threejs. https://github.com/jupyter-widgets/pythreejs/issues
Received several issue notifications in the last week, mostly centered on ipywidgets, including concerns over a soon to be released ipywidgets 8. https://github.com/jupyter-widgets/ipywidgets/issues
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Status Update.
The existing code. Up till now the total number of protons in an occupied slot (an integer from 1-6 found in the slotset_df dataframe’s slot row and ‘Protons’ column) is divided by two. The quotient becomes the number of that slot’s alphas and the odd remainder is added as a single proton (+ electron) at the end of the slot’s proton group additions. Neutrons are added last, to either slot end if needed, subjective additions to the datafame, mostly used to balance the atoms with many more protons than neutrons, whose slots contain mostly odd numbered protons and fewer alphas. Finally, all the particles in a slot spun in a single direction.
That scheme was sufficient to allow the creation of crude charge field atomic models, but it is not adequate for displaying charge field bonding between atoms. While adding contra spins within each slot was a welcome and necessary improvement, more change is needed.
The current goal. As I mentioned last time, the next task is to better define and display a slot’s internal proton stack bonding. The dataframe slot contents and program code need to be expanded to include each of that slot’s up to six proton groups. Furthermore, in the simple case, I believe that the rules governing bonds within a given slot will apply equally well to a bond between two atoms sharing a single slot.
Happy to report I think I’ve got enough information below to proceed. Your feedback is of course welcome.

From Diatomic Hydrogen *. Miles wrote.
Therefore, the right spinning proton can capture at either pole, and so can the left spinning proton.
So instead of just e-top and e-bottom, we have e-top-left, e-top-right, e-bottom-left, and
e-bottom-right. I will simplify this to TL, TR, BL, and BR. If we bring these atoms together, we now have 8 possible combinations. But only 4 of these combinations create an attraction and therefore a bond.
Miles identified four valid diatomic hydrogen configurations in his figure 1 - TLBL. TRTR. BRBR. and TRBL. I copied figure 1, added yellow/black labels to the valid columns, then rotated the diagram 180 degrees in the page plane and added a second set of labels to the 4 valid configurations again. The upside down copy is included for ease in reference and identifying each electron position (T or B) and proton spin (L or R) configuration.
Basically, an electron orbiting a proton pole creates a charge minimum at the proton’s opposite pole. Properly configured and aligned electron-proton atoms will be be charge pressured together to create an atomic bond. The four configurations are valid because of three rules:
Rule 1. Two protons, each with any spin direction (L or R) can bond when they share a T B electron position configuration.
Rule 2. T T or B B electron position configurations will be valid only if both protons have the same (L or R) spin direction.
Rule 3. No combination of B T electrons are valid, two electrons between two protons prevent bonding.
Miles mentions that others will see more combinations than the initial 8 he provided in figure 1 - as I do, but he assures us he’s showing us the essentials, given proton A above and proton B below. Myself, not quite understanding the bare minimum reasoning, nor knowing which proton A or B is up, chose to label the valid configurations from both up or down directions. When viewed upside down, 3 of the 4 configurations are of course the same, yet have different labels:
1. TRBR. 2. BLBL. 3. TLTL. 4. TRBL.
TRBL is the same upside down or not. Why is TRBL’s rotational opposite, TLBR not included in either list? Anyway, for my arbitrarily directioned, limited understanding and coding intent, I’ll argue that when the program builds a single valid diatomic hydrogen in some stack, it must be one of the following 8 types:
1. TLBL. 2. TLBR. 3. TRBL. 4. TRBR. 5. TRTR. 6. TLTL 7. BRBR. 8. BLBL.
The first four are T B configurations, valid by rule 1. The second four are T T and B B configurations, they are valid by rule 2 since both protons spin in the same direction. None of these 8 H2 configurations violate rule 3.
Miles wrote.
Notice that I have matched the known data for ortho- and para-hydrogen. We are told that equal spinning diatoms outnumber opposites by 3 to 1. Which is what I found in my mechanical combinations.
I spent some time trying to reconcile my list of 8 valid bonds with Miles’ list of 4 and settled on the “unspecified proton A or B” argument as the difference, A and B establish up/down directionality. Since the 3 to 1 ortho- and para-hydrogen ratio, now increased to 6 to 2 remains the same, I don’t see any problem in trying to code diatomic hydrogen proton bonding from the larger – two directional list of 8 valid configurations. In any case, I’ll reserve the option to convert to Miles list of four H2’s, then maybe adding a calculated or random flip and/or 180 degree rotation for variety when positioning them.
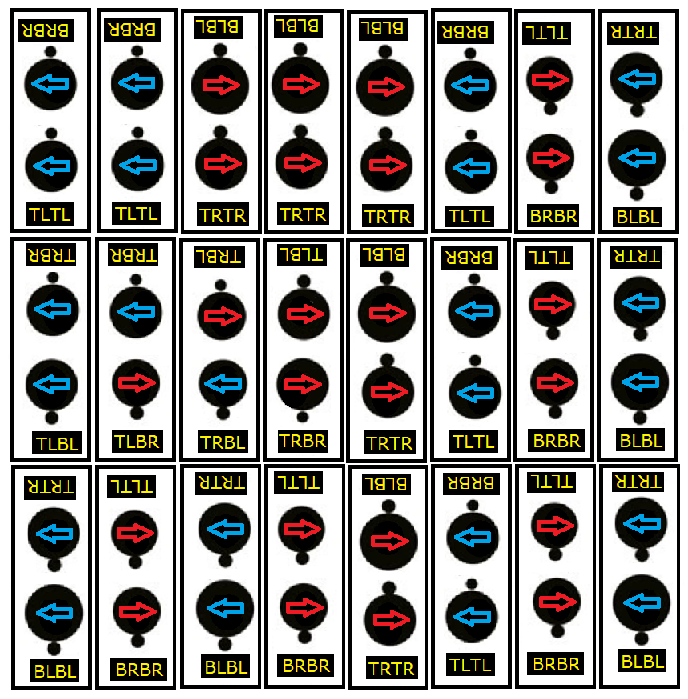
Figure 2. Following figure 1. Proton labels A and B are replaced with proton spin direction red or blue (R or L) arrows. 24 vertical black rectangles, each containing a valid H2 configuration. Eight columns of valid slot sequences for 6 proton groups (proton and electron), grouped into 3 vertically aligned H2’s.
All right, that accounts for all the valid bonds between two electron/proton H2 groups. How might a third electron-proton be added?
Let proton rows 3 and 4 comprise all 8 of the valid up/down diatomic configurations. The second row contains all allowed H2’s: 1. TLBL. 2. TLBR. 3. TRBL. 4. TRBR. 5. TRTR. 6. TLTL 7. BRBR. 8. BLBL. Upside down, rows 2 and 3 still contain all H2’s from that list, but in a different order.
I had intended to diagram single electron-proton, H additions, up and down from there, but by rule 2, adding valid electron-proton configurations to the ends of any valid H2, there’s only one answer. We can only add electron-protons with the same electron position (T T or B B) and the same proton spin. All added H types must be the same as the H type to which it is added, either BL, BR, TL or TR. The third H type added to either end will be the same as the second H type. The fourth H type will be the same as the third. The whole diagram jumps out at once, it shows all possible H2 configurations above and below that center H2.
Here’s the same data as figure 2, each row is a sequence of 3 H2’s or 6 H’s.
1. TLTL, TLBL, BLBL.
2. TLTL, TLBR, BRBR.
3. TRTR, TRBL, BLBL.
4. TRTR, TRBR, BRBR.
5. TRTR, TRTR, TRTR.
6. TLTL, TLTL, TLTL.
7. BRBR, BRBR, BRBR.
8. BLBL, BLBL, BLBL.
Moving the 4 T B configurations from fig 2 central rows (3 and 4) or listed rows (1 and 2) - then adding 2 H2’s downward; or moving fig 2 central rows (3 and 4) to rows (5 and 6) then adding two H2’s upward; shifts fig 2’s diagram up or down, and shifts these three lists left or right. As in figure 2, the overall pattern is easier to see than it is to explain. The shifts are shown below, only rows 1 – 4 are listed since rows 5 - 8 are of a single H2 type and cannot be “shifted” to form a different valid sequence.
Shift the T B H2 to the left.
1. TLBL, BLBL, BLBL.
2. TLBR, BRBR, BRBR.
3. TRBL, BLBL, BLBL
4. TRBR, BRBR, BRBR
Shift the T B H2 to the right.
1. TLTL, TLTL, TLBL.
2. TLTL, TLTL, TLBR.
3. TRTR, TRTR, TRBL.
4. TRTR, TRTR, TRBR.
What about neutrons? Don’t neutrons blow up figure 2? And alphas, do all valid H2’s form unique and valid alphas?
I haven’t given it much thought yet. For one thing, the presence of the electron at the T or B proton pole position results in a charge minimum at the other proton pole sufficient to attract and bond with a properly configured and positioned H atom. Neutrons must also be attracted to those same low charge pressure zones.
More importantly, bonding-wise, neutrons are just like protons. They spin left or right and they just as likely as protons to have an electron orbiting their top or bottom charge intake poles, along with the resulting charge pressure imbalances and bonding. It seems pretty clear that neutrons must follow the same bonding rules as protons. Electron and anti-electrons cannot share the same internal space where a bond cannot form (rule 3). In a T T or B B electron configuration - the neutron must spin in the direction as the previous proton (or neutron?) (rule 2).
Alphas are beyond me at the moment.
With respect to mBuilder code, I think I have enough to work with. Redefining the slot’s proton group contents. Some slots may indeed spin in a single direction, or slots may spin in just two directions, meeting somewhere near the center of the stack. It seems I need to add new electron+proton groups such as BL, BR, TL or TR, and perhaps also electron+proton+neutron groups, BL+N, BR+N, TL+N and TR+N. Stacks may include alternating electron-proton or electron-neutron particle groups.
*
332. Diatomic Hydrogen. http://milesmathis.com/diatom.pdf My new charge bonding explains this much better than electron sharing. Plus an analysis of spin isomers. 9pp.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
https://phys.org/news/2021-09-magic-tin-tail.html#!
Chromium6- Posts : 735
Join date : 2019-11-29
LongtimeAirman likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Thanks Cr6, If I understand correctly, this is an excellent opportunity to show off the Atom Builder (AB) program, by comparing charge field atomic models to conventional, magical atomic theory.
Thanks Lloyd.
Grabbing magic tin by the tail By Cern. An article about the difficulty and current attempts to measure the energy in the doubly magic nuclei of Tin-100, with 50 protons and 50 neutrons.
https://phys.org/news/2021-09-magic-tin-tail.html#!
For more details that article references, https://doi.org/10.1038/s41567-021-01326-9
Mass measurements of 99–101In challenge ab initio nuclear theory of the nuclide 100Sn
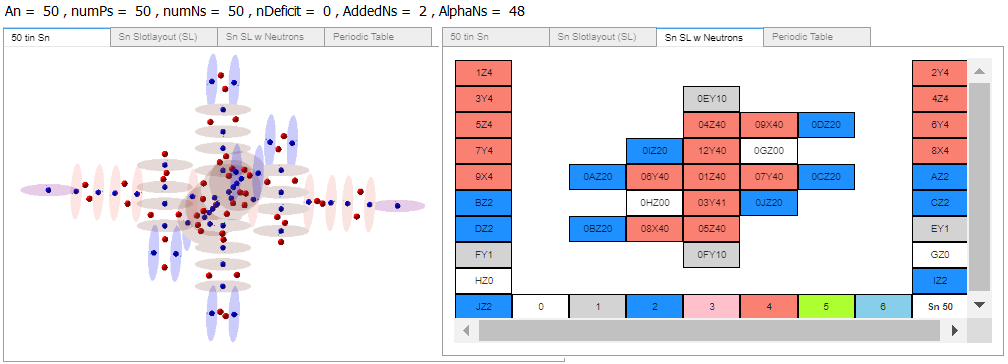
Here’s AB’s Tin-100, 100Sn, with 50 protons and 50 neutrons. The atomic image’s z axis is left right, and in the SL diagram the z-axis is up-down. A temporary in-place print addition gave me the proton neutron counts at the top.
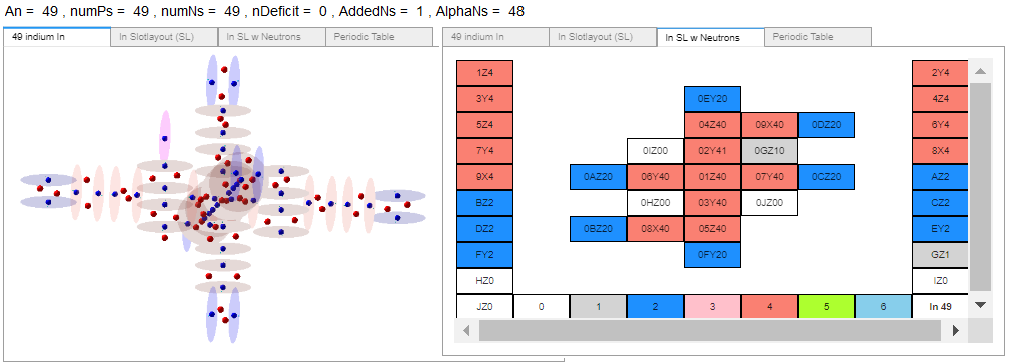
And Indium-98, 98In, with 49 protons and 49 neutrons.
Easy to compare the main proton and neutron differences between the atoms as well as imagine where neutrons might be added or subtracted.
The program only displays a single proton plus neutron atomic weight for any given atom. It wouldn’t be difficult to add atomic isotopes to the program, the hard part would be identifying all valid possible neutron positions - vacancies or additions, for each atom.
Assuming the charge field models of Tin and Indium are ‘correct’, here are the differences I see.
1a. Tn-100: contains single protons in top and bottom slots 14 and 15.
1b. In-98: contains alphas in top and bottom slots 14 and 15.
2a. Tn-100: hook slots 18 and 19 both contain alphas.
2b. In-98: hook slot 16 contains a single proton.
3a. Tn-100: has added neutrons at slots 2 and 3.
3b. In-98: has one neutron added to slot2.
Tin has more protons and neutrons concentrated near its center than Indium. Enough to suggest Indium is not as similar to Tin as the researchers believe.
To arrive at 100In I suppose I might add two neutrons to Indium’s slot 2 top and bottom, or hook slots 17 and 19. Easy enough to add two red neutron dots to the image shown. Then compare 100Sn to 100In directly.
//////\\\\\////\\\//\/\\///\\\\/////\\\\\\
More Thoughts on Bonding between Neutrons and Protons; and Alphas. Great thoughts? Not likely, they keep changing. My question is: What makes a valid configuration of protons, neutrons and elections in a given slot with 1-6 protons? I’ve been flailing about attempting to generalize from my poor understanding of alphas and diatomic hydrogens.
Last time I indicated that I’d go ahead and assume that the eight valid configurations of diatomic hydrogen (1. TLBL. 2. TLBR. 3. TRBL. 4. TRBR. 5. TRTR. 6. TLTL 7. BRBR. 8. BLBL.) vice Miles’ four (TLBL. TRTR. BRBR. and TRBL) can include two alpha neutrons. Well that must be wrong. As Miles clearly states, the neutrons within the alpha are reversed, one must spin L and the other must spin R. Plus, within a given e-p-n, assuming neutrons do not flip when forming an alpha, I believe the neutron must spin in the same direction as the proton. Then both the top and bottom 2Hs must spin in opposite directions. In which case, only two of the 8 diatomic configurations can form an alpha, (Miles would say 1) TLBR or TRBL.
I also asserted neutrons must follow the same bonding rules as protons. That is, an electron-neutron can bond to an electron-proton as long as, by rule 2: 1. both e-p and e-n were the same electron position configuration and 2. both proton/neutron spins were in the same R or L spin direction. If alphas were built up from e-p and e-n pairs, then alphas should have four electrons instead of two. Suffice to say I’m likely mistaken, but that’s not to say slots cannot be built up using e-ps and e-ns.
\//\\\////\\\\\//////\\\\\\/////\\\\///\\/
1. MIT License. Forgot to mention last time, I added an MIT license to the project repository at https://github.com/LtAirman/ChargeFieldTopics
I hope its correct. I thought the process would be more involved than just filling in a name and date using a format found on-line.
https://choosealicense.com/licenses/mit/
2. Updated the project's README file.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Looks like they are working on that error with the jupyter-client:
https://github.com/jupyter-widgets/pythreejs/issues/366
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Just wanted to get your thoughts on molecule to molecule with sodium-graphene. Looks like this company JANUS has decent research on a new battery.

https://www.science.org/doi/epdf/10.1126/sciadv.abf0812
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Sorry, no project progress to report his week, I'm in a mental loop; what makes a valid slot configuration?
/\\///\\\\///\\/\
Any thoughts or feedback on my summary Tin/Indium comparison for better understanding of “Grabbing magic tin by the tail“ by Cern last week?
\\\\///\\/\\///\\\\///\\
On the Issue #366 page, the last action shown is from 22 days ago.Cr6 wrote. Looks like they are working on that error with the jupyter-client:
https://github.com/jupyter-widgets/pythreejs/issues/366
29 days ago vidartf opened a consolidated list of issues - Issue 378 (including Issue 366) requiring attention for the upcoming ipywidgets 8 release.Vidartf wrote. linked a pull request 22 days ago that will close this issue
https://github.com/jupyter-widgets/pythreejs/pull/378
On page #378, 22 days ago, vidartf made 4 code commits including possible fixes as well as re-identifying the non-finite floats problem.vidartf wrote. Ipywidgets 8 is about to be released, and some changes are needed to support it: …
Since then, still on page #378, one new item, yesterday. Referring to Issue 366, vidartf asked @akaszynski Do you want to do a review / test this?Vidartf wrote. jupyter_client has deprecated support for non-finite floats, and aggressively prints to stderr if you try to serialize them.
Notes: We do not need a deserializer, as CFloats will handle the conversion in its validator. We were already relying on this fact, so no change. Similarly, the JS convertFloat[...] functions continue to serialize/deserialize there. This also extends Vector/Matrix/Euler to support IEEE floats.
I don’t know if there’s a fix in or not, vidartf certainly appears on top of it.
\\\\\////\\\//\//\\\////\\\\\
I downloaded Real-time imaging of Na+ reversible intercalation in “Janus” graphene stacks for battery applications and have briefly looked at it a few times. It seems sheets of graphene separated by spacer molecules including sodium can store charge or pass charge comparable to lithium ion batteries. I'm afraid I would need to devote a great deal of time and effort in order to begin to understand it. Was there anything specific that you had in mind? Why not include it on your Graphene thread?Cr6 wrote. Just wanted to get your thoughts on molecule to molecule with sodium-graphene. Looks like this company JANUS has decent research on a new battery.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
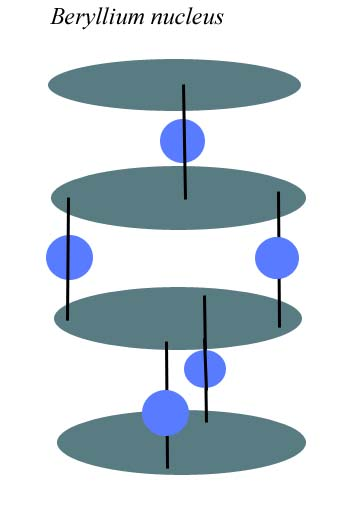
Miles’ beryllium atom.
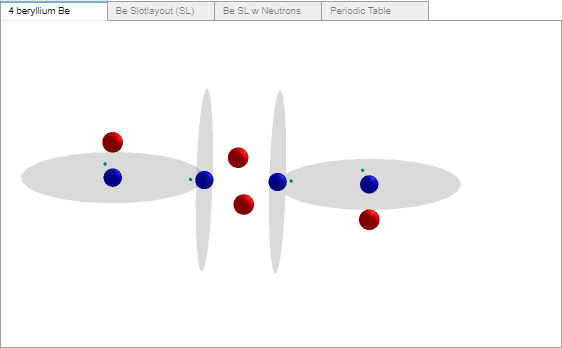
AtomBuilder2’s beryllium atom. I believe I’ve mentioned not being happy in implementing a beryllium atom so much different from Miles’ image.
Alpha Dilemma. Recap, the current program builds atomic slot proton stacks containing the odd proton and/or 1,2, or 3 alphas. A recent reading of Diatomic Hydrogen (DH) convinced me that a slot cannot contain more than a single alpha. Here’s my reasoning.
First, neutrons within an alpha have opposite axial spin directions. Note, opposite axial spins seems somewhat at odds with the fact that both neutrons must orbit the alpha’s proton1 pole to proton2 pole straight through charge channel in the same direction – while recycling charge and anti-charge vortices.
Next, by my interpretation of the DH rules(?) each neutron should have the same axial spin direction as the proton it bonded to or arrived with. The top and bottom alpha protons would then also spin in opposite directions, LR or RL. Using DH rules we can then add up to 4 e-p’s to arrive at proton slots with spins LLLRRR or RRRLLL.
I imagine two vertically adjacent alphas would break a slots’ charge channel into three sections, top, center and bottom with, say, three pairs of protons with alternating spin directions, LLRRLL or RRLLRR.
In DH, the electron and anti-electron between 2 protons prevent any bonding, well then, a third spin direction created by adjacent alphas and opposite vortices would also chop up the slot’s charge channel and prevent two alphas from bonding.
A lose rationale and plenty of doubt, please let me know if you disagree.
Bi-directional and single directional slots. Complicating matters, as I posted on 3 June, there’s no reason I can think of preventing alpha-less slots in which all the protons have the same e-p configuration and the same proton spin direction.
Neutrons. I'll assume electron-neutron (e-n) pairs follow e-p bonding rules and can be found between adjacent protons. Those neutrons will not align with, but will instead orbit the proton-proton straight through charge channel. The resulting beryllium atom will appear more similar to Miles’.
Slot1. Slot1 may be special. Sure it's the center of the atom and all atoms have an occupied slot1. Hydrogen, H, contains a single proton at slot1's center (0,0,0). Currently, I believe I’ll code all other atoms, He and above, with a single alpha at or near slot1's center (0,0,0). The alpha may be off-centered within the slot as odd numbered new e-p units are added above or below the alpha. With charge entering the alpha from below, and anti-charge entering the alpha from above, it makes sense to place the bi-directional configuration in slot 1.
Single or Bi slot spin directions(?). Likewise, if it makes sense that all protons in a given slot spin in a single direction, it would apply to the z-axis slot4 and slot5, above and below slot1, so the single directional proton spin groups might spin in the same direction as slot1’s corresponding top or bottom. X and y-axis slots 6-9 might also be possible candidates for single spin direction slots. I’ll try modeling them both for comparison.
Changes. Working to implement the new proton stack configurations.
Added a new function, buildProStackM(a,j), not yet complete, intended to replace the program’s current function, buildProtonStackN(a,j). Or perhaps allow the user to select between the alternate slot configurations.
a. Coded the six sets of z-positions (pPositions) for a slot of 2-6 protons including a central alpha.
b. Coded the six sets of z-positions for a slot of 1-6 protons without an alpha.
Sound Ok? Or am I wasting time?
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Opposite configurations. On the left, dsSlots=[0,1,2,9,10,11,12,13,14,15,16,17,18]
and ssSlots=[3,4,5,6,7,8]. Flipping the two, on the right, dsSlots=[3,4,5,6,7,8],
ssSlots=[0,1,2,9,10,11,12,13,14,15,16,17,18]. Slots 1-19 are addressed with
indices 0-18. Not all slots are occupied. The electrons are enlarged and protons
brought closer together for easier viewing.
Made some progress in developing the single vs. double proton spin directional
group configurations. buildProStackM(a,j):.
1. Added new lists: dsSlots, listing the double spin direction slots; and ssSlots,
containing the single proton spin direction slots, enumerated above. There are
right and left single proton spin direction groups sSlotsR=[0,2,10,12,14,16,18]
and sSlotsL=[1,9,11,13,15,17] apply to the right-side image.
2. Added a new slot spin assignment section to ensure correct spins.
- Code:
if j in ssSlots: # Single spin slot spin assignments.
if j in sSlotsR:
stacksA[j].add(protonMi)
else:
stacksB[j].add(protonMi)
else : # Double spin slot spin assignments.
if num%2 != 0:
stacksA[j].add(protonMi)
else:
stacksB[j].add(protonMi)
3. Added code to specify electron positions, above or below the protons.
The new slots appear simpler, non-alpha neutrons have yet to be added, but the
new configurations are more stringent with respect to proton spin directions
(R or L) and electron positions (T or B).
My initial assumption of an alpha in the center of a double spinning set of protons
in slot1, as shown on the left-side image, at the center of the carousel, now strikes
me as wrong. Hard to tell from the image but the neutrons in an alpha centered at
(0,0,0) orbit about the +/- z-axis while also passing directly through the front/back,
and left/right (+/-x and +/-y) charge channel currents. That would result in a great
deal of alpha interference. Strike that idea. Having the model allowed me to simply
swap my lists of single and double spinning slots, thereby creating the right side
image which does not suffer any direct side channel, “alpha interference”. That
doesn’t mean it’s right, but there’s no reason to assume its wrong.
Next I’ll need to start adding neutrons.
P.S. Correcting: 1. Changing "buildProtonStackN(a,j)" to "buildProStackM(a,j)".
2. Added "for easier" before "viewing."
.
Last edited by LongtimeAirman on Fri Jul 01, 2022 8:33 pm; edited 1 time in total (Reason for editing : Added P.S.)
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Bad news,
The three project notebooks: AtomBuilder, AtomBuilder2 and mBuilder no longer display a pythreejs rendered output.
I left the following message at both locations:
https://github.com/jupyter-widgets/pythreejs/pull/378
https://github.com/jupyter-widgets/pythreejs/issues/366
Hello vidartf,
I’m a newbie programmer working with Jupyter Notebook and pythreejs.
All 3 of my project files at https://github.com/LtAirman/ChargeFieldTopics stopped working this morning. They no longer display the pythreejs rendered scene.
Over the last few months the cell setting up the rendered scene displayed a UserWarning, due to non JSON compliant infinite values. I believe you identified the problem, involving the pythreejs orbital camera, in issues 366 and 378 which I’ve been following.
It was my impression this non-finite number problem would be corrected with the upcoming ipywidget 8 release, seeing no rendered output at all is an awful surprise. What do I do now?
I’d appreciate any guidance or information you might provide in this matter.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
LongtimeAirman wrote:.
Bad news,
The three project notebooks: AtomBuilder, AtomBuilder2 and mBuilder no longer display a pythreejs rendered output.
I left the following message at both locations:
https://github.com/jupyter-widgets/pythreejs/pull/378
https://github.com/jupyter-widgets/pythreejs/issues/366
Hello vidartf,
I’m a newbie programmer working with Jupyter Notebook and pythreejs.
All 3 of my project files at https://github.com/LtAirman/ChargeFieldTopics stopped working this morning. They no longer display the pythreejs rendered scene.
Over the last few months the cell setting up the rendered scene displayed a UserWarning, due to non JSON compliant infinite values. I believe you identified the problem, involving the pythreejs orbital camera, in issues 366 and 378 which I’ve been following.
It was my impression this non-finite number problem would be corrected with the upcoming ipywidget 8 release, seeing no rendered output at all is an awful surprise. What do I do now?
I’d appreciate any guidance or information you might provide in this matter.
.
Ouch. This is really sad news if they don't fix this bug.
This might be an option for a port if necessary:
https://www.ogre3d.org/
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
The atoms were back again this morning Cr6. I left another message (or two) at the same locations. At #366,
Update, happy to report my projects' pythreejs rendered outputs are now working properly. Along with the UserWarning that is. I double posted this message at #378.
My day usually starts with launching Anaconda. Anaconda then loads all the packages (almost 300) for the project’s environment, which enables good version control and things like that. Next, in that environment I launch Jupyter Notebook and then open the project file. Yesterday was the first time in almost a year when there was no rendered display. I looked at my other ‘older’ pythreejs examples, they all had blank outputs and the same UserWarning. I thought the worst and left vidartf the message I posted yesterday.
I don't know what happened. Or whether there was any corrective action or not. Maybe yesterday’s problem was unrelated to #366 (the pythreejs orbital camera values) and could have been corrected with an Anaconda shut-down and restart. I've never seen any indication that would be necessary, but I’ll try it next time if it happens again. What’s the proper procedure to report any problems anyway?
I delayed posting the Good news, hoping that vidartf would reply.
Pardon my alarm, other than my exceedingly slow progress I think we’re Ok. I’ll put off investigating Ogre3D for the time being.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

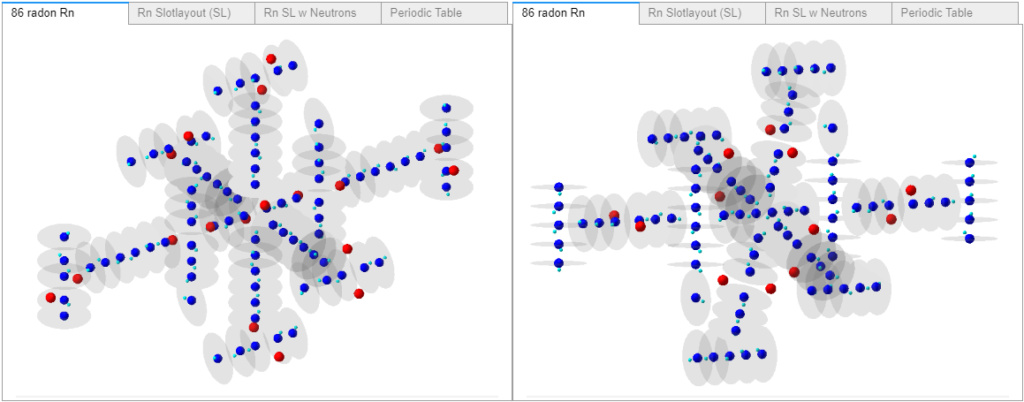
Radon, now with plenty of neutrons - some will need to be removed and/or reconfigured. Following the same new slotlayout proton stack configuration as the right-side image I posted on 1 July.
Some progress.
1. Added neutrons to the new buildProStackM(a,j) function. As shown in the Radon image above.
I’m finally past my mental loop - what makes a valid slot proton stack configuration? Recently, I asserted to the effect: electron-neutrons should follow the same bonding rules as electron-protons, for the same reasons; so a given alpha should have four electrons, 2 at the T and B proton poles and 2 at the T T or B B neutron poles.
Well, making models requires making some assumptions at times, I now consider that one wrong. Thinking about the fact that a proton’s +/- equatorial emissions likely help prevent electrons (and neutrons) from escaping a protons’ pole. The neutron’s lack of +/- equatorial emissions make electrons orbiting a neutron pole more exposed and therefore less stable than electrons orbiting a proton’s pole. Electrons are more easily knocked free of the less powerful (0.68 that of a proton) neutron pole input charge vortices. When and if electron-neutron pairs form, they may not last long. Given that rational, I no longer think it’s necessary to add new neutron dedicated electrons. For the new buildProStackM(a,j) function, all electrons only orbit proton poles and not neutron poles. With that, I was able to resume progress.
Proton groups. In the code, the 0-6 protons in a given slot are added one at a time. Each proton is part of a proton group usually including a neutron and an electron. Here’s an exhaustive list of all the proton groups I’m currently coding.
a. Electron-Proton. Used for the first proton of an alpha. Also used for neutron-less center slot positions of single proton spin groups through which X, Y or Z charge channels pass. I‘ll assume direct charge channels prevent neutrons from occupying that direct charge channel exposed slot center position.
b. Electron-2 Neutrons-Proton. Used for the second proton group of an alpha, to allow both neutrons to orbit about the alpha’s direct charge channel in the same direction, with respect to the second proton. Two orbiting neutrons can also be placed between non-alpha adjacent protons.
c. Electron-Neutron-Proton. The main proton group assigned to most all remaining proton slots.
d. 1 Neutron.
e. 2 Neutrons. Beside being completely empty, the 0 value in 0-6 protons per slot can also apply to d and e, just 1 or 2 neutrons. ‘Neutrons only’ may occasionally be found in the hook slots 16-19. Generally, neutrons will orbit about a charge channel, which is simulated in the code by neutrons spinning along with the proton and electron as single group about the proton group’s spin axis, usually through the proton.
I think I like it, but it will take some modifications and effort getting used to the new slot proton stack configurations. With all the rotations turned on there’s plenty of motion and no shortage of neutrons. I believe they are more “accurate” than the multiple slot alpha configurations.
2. New datafile Cr6-Elements-M.csv. As I just mentioned, mBuilder’s new atom building function will require some small modifications to the datafile. AB2 uses Cr6-Elements.csv so I’ll leave it alone. Copy created Cr6-Elements-M.csv.
a. One needed mod, on 24 June I showed that Miles depicts Beryllium as a single slot1 with a 4 proton stack and no alphas. AB2 shows the Be atom to be comprised of three occupied slots, 1-3, with slot1 containing an alpha. Be will be changed to a single slot1 proton stack in Cr6-Elements-M.csv.
b. Another mod will be to identify/remove all neutrons passing directly through +/-X, +/-Y, or +/-Z charge channels. I’ll need to review each atom on a case by case basis. I may assume atoms will have an alpha in slot1 (0,0,0) with neutrons orbiting the alpha’s z axis, as long as that atom doesn’t include +/-x or +/-y occupied slots and side charge channels.
c. I see single neutrons orbiting the single protons’ direct charge channel in the hook positions, slots 18 and 19. Might those single neutrons share the same direct charge with the proton as I’ve modeled them before? I’ll decide at some point.
d. I see abutting neutrons at adjacent slot ends, one or the other neutron might need to be removed.
e. By default, the buildProStackM(a,j) function adds plenty of neutrons. The datafile’s neutron columns may need changing.
These and likely more details requiring datafile changes makes the idea of showing alternate (old and new) proton stack layout configurations difficult and unlikely. After all is said and done I’ll probably have ended up replacing buildProtonStackN(a,j) with buildProStackM(a,j) in both AB2 and mBuilder and swapped out the old datafile.
No pythreejs rendered output #381. Three days ago I submitted the problem at github as an issue.
https://github.com/jupyter-widgets/pythreejs/issues
https://github.com/jupyter-widgets/pythreejs/issues/381
I made the following post earlier today.
8 July. This morning, after starting everything, I saw a good rendered output. I made selection changes and the rendering was still good. I went to another chrome tab, then came back to the notebook tab and the pythreejs rendering was gone.
Closed Notebook, Anaconda Navigator and chrome and restarted; no change - no renderings.
Shut down and restarted everything including my computer and the rendering was again working properly. I’ve made several different, most complex object selections without a problem. Opening, closing and minimizing tabs and using other apps and browsers like I normally do. After three four hours the pythreejs renderings are working properly.
If all I need to do is reboot my computer I suppose I can live with the problem. I’ll try logging my activities for a few days before posting again.
No reply yet, nor do I expect any at this point, I'm a little discouraged. I just checked, mBuilder, is still properly rendering in the background.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
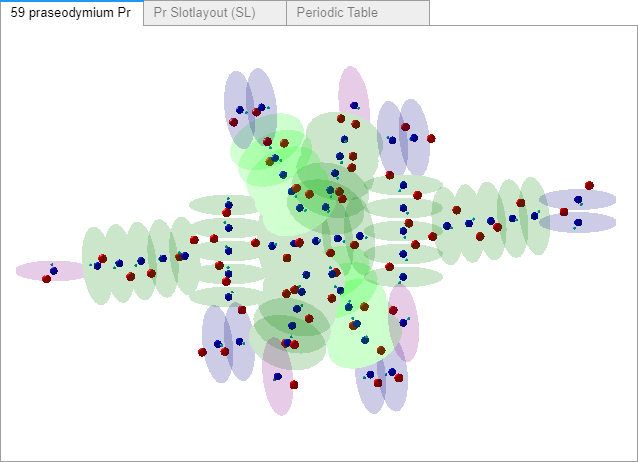
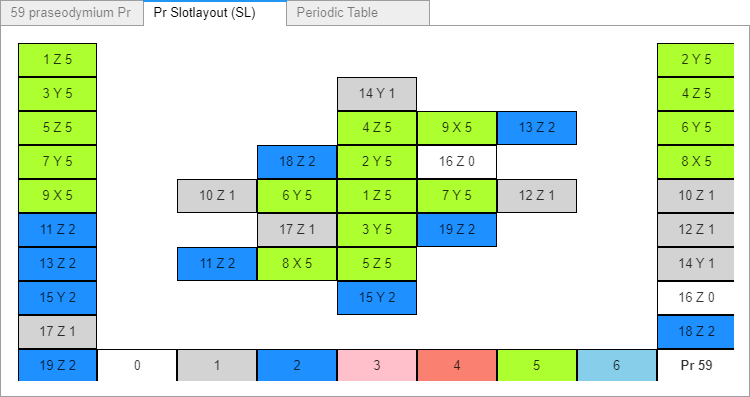
The Praseodymium charge field atom shown here doesn’t include any alphas or two directional proton spin slots – yet. All slots are filled with single proton spin directional groups (either Right or Left), and single (Top or Bottom) electron configurations. Electrons and Neutrons in the 1-6 proton groups in every slot are on opposite sides of the proton. Four possible configurations I've tried to apply logically.
With difficulties and further consideration, and, I apologize - further delay, I decided to initially load all slots as single proton direction spin groups and electron configurations. I’ll add alphas separately.
Changes.
Previous: Neutrons NL NR N2N3 N12 NE NZ AlphaType SlotOrien SlotSpin
Current: Neutrons N1 N2 N3 N4 N5 N6 Electrons SlotOrien SlotSpin
1. The Cr6-Elements-M.csv datafile contained 4 unused neutron location data columns (N2N3, N12, NE, NZ). NL and NR were used to add neutrons to the slot’s R and L ends, and also by the SlotLayout with Neutrons diagram found in AB2 tab 3. Since neutrons are being added somewhat differently in this new slot configuration build function I decided to throw out that diagram, easy enough to rebuild again later if desired. Those 2+4 data columns are now changed to account for the 1-6 slot neutron counts (N1-N6), each with possible integer values 0,1 or 2. The (N1-N6) data has not yet been entered or accessed by the buildProStackM() function yet.
2. Converted the “AlphaType” column (From how many alphas in that slot, 0-3) into “Electrons”, indicating that slot’s electron configuration, either: Top, Bottom, None (for empty slots) or Alpha (both T and B). All 90 atoms’ 19 slots’ “Electrons” column are currently filled with T’s, B’s, and N’s. Except for He He, Li and Be slot1 “Electrons” data which are A’s.
3. “SlotSpin” was also unused. It now contains proton spin directional values Right, Left, or None (again empty slots are filled with N’s), or A (an alpha’s protons spin in both R and L spin directions). The datafile’s “SlotSpin” column is filled with R’s, L’s, and N’s. Except for He, Li and Be slot1 “SlotSpin” data values are also A’s.
No pythreejs rendered output #381. There has been No problem since my last post.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

All atoms are reconfigured to build the new slot configurations as read from the latest data file changes.
Thanks Cr6, at the moment, between you and me, my objectivity is shot and I’m delirious.
Implementing slot particle configurations based on proton spin directions (R or L) and electron at proton (T or B) pole as suggested in Diatomic Hydrogen makes a whole lot more sense then building slots from alphas and the odd proton. Easier to read and make changes to the electron, neutron and proton particle positions directly in the data file, also an improvement. And Neutrons! There are plenty of neutrons and plenty of positions they might occupy. Sorry it took me so long, but incremental improvements are better than none.
Given some of the possible new alternatives I’ve made a few choices.
Beryllium, Be. Above. The two central protons, neutrons and electrons form a TLBR alpha, a two proton spin directional slot group. The top two electrons, protons and neutrons spin L, while the bottom 3 neutrons, 2 protons and 2 electrons spin R. Be’s slot1 datafile neutron configuration is:
Neutrons 5 N1 0 N2 2 N3 1 N4 2 N5 0 N6 0 Electrons A SlotOrien Z SlotSpin A.
A little confusing, the slot is built +/-z starting with the center alpha – the second and third protons from the top. Zero neutrons are added to the first L proton and two neutrons are added to the second R proton. L Proton 3 is at the top, and R proton 4 is at the bottom. The neutrons are added at the same time and in the same order. Compare to Miles’ Be nucleus I posted on 24 June. As I recall, Miles didn’t indicate whether an alpha is included in Be or not. The same neutron configuration can be included in a single directional proton spin group, R or L, but then all electrons would need to be a single electron configuration, T or B, and not both T and B as shown here.
Lead, Pb. Below. Found/corrected a slot11 x position error. Look closely and you’ll see a gap between slot1’s two central protons. The electrons are present but on the opposite proton poles - in an alpha T B configuration – minus the neutrons(!). While it makes sense to me to remove any neutrons from the direct +/- x, y, side channels, its just a guess on my part and I’m often wrong. Another choice, all Pb proton stacks containing six protons have five intervals which contain six neutrons. One of the five intervals has two neutrons. I did that because I didn’t like hanging a neutron off of a slot’s end, preferring to leave both slot ends as protons (not to mention the electrons). In other elements with large stacks I left those neutrons orbiting outside the outermost protons and chose not to include neutron doubling. Likewise, sometimes I included single neutrons with single protons, as with Hydrogen, but generally not as with Pb’s hook position protons. More alternatives later.
Ok, the display neutrons checkbox is again working properly.
Next, code review – again – and cleanup.
No pythreejs rendered output #381? Another week with No problem.

.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
With a rules engine and big data at this level it might be possible to really make "bonds" happen for molecules. I may need to database this again.
Chromium6- Posts : 735
Join date : 2019-11-29
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
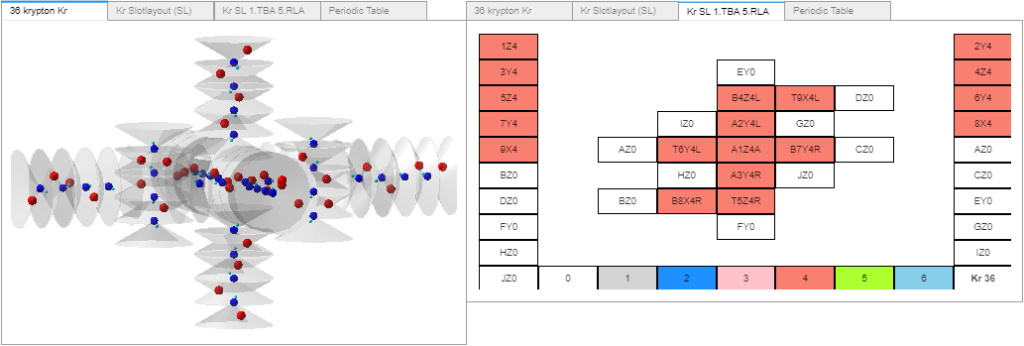
Krypton, now described by a new slot layout diagram, 'SL 1.TBA 5.RLA'. Unfortunately, neutrons are not included.
Last week I’d implemented the 2-directional slot1 TLBR, as well as distributing all the four possible single-spin groups (created from multiples of TR’s, TL’s, BR’s and BL’s) throughout all the remaining slots. Since then, I’ve restored all the alphas in slot1, reserving the right to mess with them again in the future. Yesterday I implemented the single spin directional slots with TB electron positions: TLBL (in slot2) and TRBR (in slot3). I still need to add a TRBL.
'SL 1.TBA 5.RLA'. In the meantime I made a second SL diagram to include the new data. Occupied slots in the center SL contain 5 digits: 1.TBA. Electron position at the proton's T, B or A pole. Note that A can only indicate TB since a BT electron configuration is not allowed. 2. The slot number (1-19), as a single digit, 1-J. 3. The slot's proton emission plane orientation. 4. The color-coded number of protons in that slot. 5.RLA. The slot's proton groups’ spin direction. An A in the fifth position indicates a top of the slot proton spin of L or R and the bottom of the proton slot spin of R or L, I’ll probably need a new fifth position single digit besides A (i.e. applicable to slot1’s fifth digit) to differentiate between LR or RL.
Again, slot1 is a TLBR, slot2 is a TLBL and slot3 is a TRBR. All other slots are TRTR…, TLTL..., BRBR..., or BLBL…. I’m not at all sure how to assign specific configuration types to specific slots. All in all, including the slots’ electron position and proton spin data may be a bit more complicated, I’d say the resulting model is definitely improved.
I did a couple of searches and read about “rules engine and big data“. I agree, we now have a few specific rules for bonding proton groups within a single slot’s stack. Atomic bonding may be slightly different – between stacks(?). Clear or not, rules must first be identified. Bear in mind that a rules engine can be extremely complicated and even counterproductive in the long run, whereas an if/then/else logic structure or table might suffice. At least for starters. For example, here’s the current buildProStackM(a,j) function. Note the if/then/else logic structure beginning halfway through, used to build each new type slot configuration.Cr6 wrote. With a rules engine and big data at this level it might be possible to really make "bonds" happen for molecules. I may need to database this again.
- Code:
def buildProStackM(a,j):
"""
Parameters: a - total number of slot protons. j - given slot #.
buildProStackM(a,j) is an attempt to follow Miles Mathis'
paper,'Diatomic Hydrogen' (DH), which includes electron-proton
(e-p) to e-p charge bonding summarized in rules 1, 2, and 3:
Rule 1. Two protons, each with any spin direction (L or R) can
bond when they share a T B electron position configuration.
Rule 2. T T or B B electron position configurations will be valid
only if both protons have the same (L or R) spin direction.
Rule 3. No combination of B T electrons are valid, two electrons
between two protons prevent bonding.
"""
if coloredEmissions.value == True:
for k in range(6):
if a == k + 1 :
emissionmaterial = emissionMaterialList[k]
else:
emissionmaterial = emissionMaterialList[6]
"""
p2p3 = 2*p2p3Dist*pRadius, distance added for each additional e-p.
The six sets of equidistant proton positions for a slot with 1-6 protons.
The proton (groups) are added beginning at the slot center. 5,3,1,2,4,6.
"""
if a == 1 : pPositions = [0]
if a == 2 : pPositions = [p2p3/2,-p2p3/2]
if a == 3 : pPositions = [0,-p2p3, p2p3]
if a == 4 : pPositions = [p2p3/2,-p2p3/2,3*p2p3/2,-3*p2p3/2]
if a == 5 : pPositions = [0,-p2p3,p2p3,-2*p2p3,2*p2p3]
if a == 6 : pPositions = [p2p3/2,-p2p3/2,3*p2p3/2,-3*p2p3/2,5*p2p3/2,-5*p2p3/2]
def conesPlusPlane(protonMi,emission1,emission2,position):
emission1.add(Mesh(emissionGeometry, emissionmaterial))
#emission1.position = [0,0,coneHeight/2]
emission1.position = [0,0,position]
emission1.rotateX(-piG/2)
protonMi.add(emission1)
emission2.add(Mesh(emissionGeometry, emissionmaterial))
#emission2.position = [0,0,-coneHeight/2]
emission2.position = [0,0,-position]
emission2.rotateX(piG/2)
protonMi.add(emission2)
protonMi.add(Mesh(emissionGeometryP, emissionmaterial))
return
def addElectron(protonMi,electronMi,position):
electronMi.add(Mesh(electronGeometry, electronMaterial))
electronMi.position = [0,-n1n2/10,position]
electronMi.rotateZ(random()*2*piG)
protonMi.add(electronMi)
return
def singleNeutron(protonMi,neutron,position):
neutron.add(Mesh(neutronGeometry, neutronMaterial))
neutron.position = [0,-0.75*n1n2/2,position]
neutron.rotateZ(random()*2*piG)
if displayNeutrons is True :
protonMi.add(neutron)
return
def pairOfNeutrons(protonMi,neutron1,neutron2,position):
neutron1.add(Mesh(neutronGeometry, neutronMaterial))
neutron1.position = [0,-0.75*n1n2/2,0]
neutron2.add(Mesh(neutronGeometry, neutronMaterial))
neutron2.position = [0,0.75*n1n2/2,0]
neutronPair.add(neutron1)
neutronPair.add(neutron2)
neutronPair.rotateZ(random()*2*piG)
neutronPair.position = [0,0,position]
if displayNeutrons is True :
protonMi.add(neutronPair)
return
num = 0 # Increments the occupied slot's 1-6 proton groups.
while num < a:
protonMi = Group()
protonMi.add(Mesh(protonGeometry, protonMaterial))
if emissionType.value == 'plane':
protonMi.add(Mesh(emissionGeometry, emissionmaterial))
if emissionType.value == 'cones':
emission1 = Object3D()
emission2 = Object3D()
conesPlusPlane(protonMi,emission1,emission2,coneHeight/2)
# Electron and neutron additions to proton spin groups.
electronMi = Object3D()
neutronMi = Object3D()
neutronNi = Object3D()
neutronPair = Group()
# Building the various T,B,A, and R,L,A slot configurations.
intColumn = 8+num # Pointing to the N1-N6 data locations.
intValue = slotsetA_df.iloc[j,intColumn]
if slotsetA_df.loc[j+1,('SlotSpin')] == "R" or "L" :
if slotsetA_df.loc[j+1,('Electrons')] == "T" :
addElectron(protonMi,electronMi,p1e1)
if intValue == 1 :
singleNeutron(protonMi,neutronMi,-p2p3/2)
elif intValue == 2 :
pairOfNeutrons(protonMi,neutronMi,neutronNi,-p2p3/2)
if slotsetA_df.loc[j+1,('Electrons')] == "B" :
addElectron(protonMi,electronMi,-p1e1)
if intValue == 1 :
singleNeutron(protonMi,neutronMi,p2p3/2)
elif intValue == 2 :
pairOfNeutrons(protonMi,neutronMi,neutronNi,p2p3/2)
if slotsetA_df.loc[j+1,('Electrons')] == "A" : # 'SlotSpin' = R or L
if num%2 != 0:
addElectron(protonMi,electronMi,-p1e1)
if intValue == 1 :
singleNeutron(protonMi,neutronMi,p2p3/2)
elif intValue == 2 :
pairOfNeutrons(protonMi,neutronMi,neutronNi,p2p3/2)
elif num%2 == 0:
addElectron(protonMi,electronMi,p1e1)
if intValue == 1 :
singleNeutron(protonMi,neutronMi,-p2p3/2)
elif intValue == 2 :
pairOfNeutrons(protonMi,neutronMi,neutronNi,-p2p3/2)
if slotsetA_df.loc[j+1,('SlotSpin')] == "A" and slotsetA_df.loc[j+1,('Electrons')] == "A":
if num == 0 :
addElectron(protonMi,electronMi,p1e1)
if intValue == 1 :
singleNeutron(protonMi,neutronMi,p2p3/2)
if num == 1 :
if intValue == 0 :
addElectron(protonMi,electronMi,-p1e1)
if intValue == 1 :
addElectron(protonMi,electronMi,p1e1)
singleNeutron(protonMi,neutronMi,-p2p3/2)
if intValue == 2 :
addElectron(protonMi,electronMi,p1e1)
pairOfNeutrons(protonMi,neutronMi,neutronNi,-p2p3/2)
protonMi.rotateX(piG)
protonMi.rotateZ(random()*2*piG)
if num > 1 and num%2 == 0:
addElectron(protonMi,electronMi,p1e1)
if intValue == 1 :
singleNeutron(protonMi,neutronMi,-p2p3/2)
elif intValue == 2 :
pairOfNeutrons(protonMi,neutronMi,neutronNi,-p2p3/2)
if num > 1 and num%2 != 0:
if intValue == 1 :
addElectron(protonMi,electronMi,-p1e1)
singleNeutron(protonMi,neutronMi,p2p3/2)
elif intValue == 2 :
addElectron(protonMi,electronMi,-p1e1)
pairOfNeutrons(protonMi,neutronMi,neutronNi,p2p3/2)
protonMi.rotateZ(random()*2*piG)
protonMi.position = [0,0,pPositions[num]]
# Slot spin assignments
if slotsetA_df.loc[j+1,('SlotSpin')] == "R":
stacksA[j].add(protonMi)
elif slotsetA_df.loc[j+1,('SlotSpin')] == "L" :
stacksB[j].add(protonMi)
elif slotsetA_df.loc[j+1,('SlotSpin')] == "A" :
# Double spin slot spin assignments.
if num%2 != 0:
stacksA[j].add(protonMi)
else:
stacksB[j].add(protonMi)
num += 1
return
I’m extremely curious how you might “database this again”?

.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Not much code progress to report this week. Added what I believe is the final (e-p)(e-p) proton stack configuration for an alpha bond, TRBL. Doing it last made it the easiest, in about a minute or so. The details follow. Near the end of the buildProStackM(a,j) function code I posted last time there’s “elif slotsetS … == ‘A’” which tests whether that slot is supposed to be the alpha TLBR, and if true, assigns the top proton L spin and bottom proton R spin. stacksA[] are R spinners and stacksB[] are L spinners. I copied and added the elif code just above num += 1, changing ‘A’ to ‘B’, testing for TRBL new ‘SlotSpin’ code designation B, (R,L,A,B), and if true, assigns the +z proton an R spin, and the -z proton an L spin.
- Code:
elif slotsetA_df.loc[j+1,('SlotSpin')] == "B" :
# Double spin slot spin assignments.
if num%2 != 0:
stacksB[j].add(protonMi)
else:
stacksA[j].add(protonMi)
Then changed the tab name from ‘SL 1.TBA 5.RLA' to ‘SL 1.TBA 5.RLAB'. The name is intended as a brief explanation, I may change it again.
The only other ‘change’ made was to add an as of yet unused control pane dropdown choice between displaying the atom’s main, up-down z-axis charge channel either horizontally or vertically.
- Code:
zDisplayOrient = widgets.Dropdown(
options=['horizontal', 'perspective'],
value='horizontal',
description='Z Axis:',
)
I’m sure the alternative z orientation would be appreciated. As you know, from the start I’ve made the z-axis horizontal, since tall atoms fit better sideways in my short and wide display window. I’ve had plenty of practice orienting myself. Whatever the viewpoint is, briefly turning on (and off) the carousel’s (or slot1’s TLBR -z end) R spin. With the z-axis held sideways, and the closest point of the R spinning carousel spinning straight up in front of my nose, then by the right-hand rule, positive Z is towards my left. Positive X is toward my face; and positive Y is straight up.
There seems to be odd shadows in both the plane and cone emissions. I don’t know where the flip(?) is coming from.
I noticed that the control panel particle distances based on alphas need to be converted to e-p. I’ll soon hop to it.
Most of my week was spent trying to understand the new proton stacks as e-p bond configurations, actually e-p-n bond configurations.
I’d like to figure out whether there are any particular configuration constraints, such as, when assigning configuration types to each of the SlotLayout (SL) diagram’s 19 slots. Making it an e-p based SL diagram. I realize that would be a good enough reason to consider re-databasing things. Am I free to add or subtract neutrons? I’d hope to glean what makes e-p slot stacks bond together at right antgles. I don’t expect any “answers”, but will truthfully say I’ve spent plenty of time thinking about it and will no doubt continue to do so. Theory is generally beyond me. Slow methodical plodding is more my forte. Reviewing and describing things as carefully as I can, … . The usual caveats. Feel free to add.
e-p Types. All atomic protons are accompanied by electrons. The electron-proton pair is designated e-p. All protons can spin left or right (L,R), and the electron may be orbiting the proton’s top or bottom pole (T,B) so a single e-p can be of type: TL, TR, BL, or BR. The presence of the electron over the proton’s top or bottom pole creates a charge field minima outside the proton’s opposite pole. That charge imbalance allows bonding between two e-p’s, as Miles explains in Diatomic Hydrogen (DH). I went and numbered them. Rule 1. Two protons, each with any spin direction (L or R) can bond when they share a T B electron position configuration. Rule 2. T T or B B electron position configurations will be valid only if both protons have the same (L or R) spin direction. Rule 3. No combination of B T electrons are valid, opposing and interfering vortices between electron/positron pairs between adjacent proton poles prevent the protons from bonding. The present discussion and code’s progress is based on the assumption the same bonding rules also apply equally well to (e-p-n)(e-p-n) bonding within proton stacks.
I believe the following are all 8 possible e-p TB RL slot configuration types the program can now create. Each type may come in any one of five, 2–6, e-p versions.
8 possible (e-p)/(e-p) bond types and each type’s 2-6 (e-p) proton configurations.
a. TLBR, TLTLBR, TLTLBRBR, TLTLTLBRBR, TLTLTLBRBRBR.
b. TRBL, TRTRBL, TRTRB LBL, TRTRTRBLBL TRTRTRBLBLBL.
c. TLBL, TLTLBL, TLTLBLBL, TLTLTLBLBL, TLTLTLBLBLBL.
d. TRBR, TRTRBR, TRTRBRBR, TRTRTRBRBR, TRTRTRBRBRBR.
e. TRTR, TRTRTR, TRTRTRTR, TRTRTRTRTR, TRTRTRTRTRTR.
f. TLTL, TLTLTL, TLTLTLTL, TLTLTLTLTL, TLTLTLTLTLTL.
g. BRBR, BRBRBR, BRBRBRBR, BRBRBRBRBR, BRBRBRBRBRBR.
h. BLBL, BLBLBL, BLBLBLBL, BLBLBLBLBL, BLBLBLBLBLBL.
A total of 8x5, 40 stacks to choose from. Flip them all upside down and the would be labeled the same. Starting from the center of the slot, the program adds each new odd/even, e-p proton group to the +,-,+,-,+,- slot stack ends. Randomizing 50/50 e-p additions to either slot ends would add a little variety to that list.
Summary descriptions of the 8 e-p types‘ slot configurations.
a. and b. are two, 2-directional proton spin groups. All other e-p types are one directional proton group spinners.
a. thru d. are the TB electron configurations. The remaining e-p types are either T’s or B’s.
e. and f. are top single direction spinners.
g. and h. are bottom single direction spinners.
Neutrons. I keep saying I’m just following the (e-p)(e-p) bonding rules identified in Miles’ DH paper. True; but this program is also adding neutrons, e-p-n’s instead of e-p’s. A single e-p-n may include a single electron, a single proton, and 0, 1 or 2 neutrons. All the same TL, TR, BL, or BR (e-p) rules apply, with the understanding that the neutron occupies the proton pole opposite the electron. The neutron is occupying/orbiting the charge minima near the proton pole caused by the electron. The neutron circles the proton’s direct pole-to-pole charge channel far enough away so as not to prevent the addition of another e-p-n proton group along the proton pole to proton pole bond position ine. I recall Miles describing offset neutrons between vertically aligned, horizontally emitting protons as a sort of gap stabilizer which helps keep the protons separated and aligned in a stack. I couldn’t quite see how photon collisions did that, till the cone emissions make it somewhat clearer to me. Neutrons can channel 0.68 the charge a proton can (1.00). Being offset, the neutrons avoid the full strength 6 protons charge channel. Those neutrons must block some charge, so maybe these offset neutrons, circling the direct proton pole channels add to the proton stack’s charge minima and increase a stack’s bond strength.
At the same time, I assume that additional neutrons or electrons are free to come and go without disturbing the SL structure, or changing any proton stack spins or positions. Of course there’s probably an order to neutron additions and removals, energy level, etc.
The SL structure. In this e-p-n context, I suppose the SL requires a brief re-introduction. An atom’s main +/- z charge channel aligns to the main upward charge flow from the Earth, as well as downward anticharge from the Sun. The +/- x and +/- y atomic charge channels divert some of the vertical charge and anticharge received into the orthogonal, horizontal z plane in which slots along the +/- x and +/- y charge channel arms spin as a single group around slot 1, the Carousel. Hook slot positions: 16,17,18, and 19, are joined to the main up/down column at slots, 2 and 3.
Alternating in-line and orthogonal proton stacks. The SL structure is comprised of alternating proton stacks, either aligned or perpendicular (crossed or ortho) with the main atomic charge channel axis the proton stack is centered on. The main vertical z-axis channel slots (from + to - z) are (14,4,2,1,3,5,15), with ortho slots: (14,2,3,15) and in-line slots: (4,1,5). The front/back x-axis channel slots are (11,8,1,9,13), of which (8,9) are in-line, and (11,1,13) are ortho. The left/right y-axis channel slots are (10,6,1,7,12), with ortho slots (10,1,12) and in-line slots (6,7).Slot1 is aligned with z but orthogonal to both x and y charge channel axii. Slots 16-19 are ortho to slots 2-3. The ortho slots outnumber the in-line slots.
Why are most of the slots’ pole-to-pole charge channels perpendicular to earth’s charge channels? According to my charge field understanding, a proton high up in earth’s atmosphere will, over some relatively short amount of time, align its poles to the earth, and will usually become an R spinner - over the N.Hemisphere. Furthermore, the proton pole facing the earth receives more direct charge current, including more electrons. That makes the earth aligned e-p’s mostly BR types, or BL over the S Hemisphere. Helium, the alpha, is usually depicted as a two e-p vertical stack with horizontal emissions. Slot1’s proton stack alignment to the earth’s charge makes sense, I would expect all individual proton stacks of 2-6 protons within the atmosphere will align vertically to the earth below. L spinners at the +z top of the stack or R spinners at the -z bottom of the stack. I would guess that TLBR alpha type bond in the middle of slot1 would allow charge to recycle most efficiently.
Individual protons may of course be positioned perpendicularly, with earth in their equatorial emission planes. I suppose such a proton may remain in that position indefinitely, or until a few energetic collisions knock if slightly askew, and the proton emissions no longer point directly to the earth. Earth’s emissions will then begin to quickly push the proton into the pole-toward-earth, horizon emission orientation. Cleary, there must be a bond between an aligned slot1, and an adjacent perpendicular slot2 of sufficient strength to prevent both slot2 from turning out of the perpendicular position or slot1 from turning away from its in-line axis z orientation. Both Carbon (6) and Oxygen (
That's all I got!
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
The current AtomBuilder3 graphical user interface control panel.
Things are moving along albeit slowly.
Worked on the gui control panel. As I'd intended last week, the graphical user interface control panel has been cleaned-up and updated to the latest e-p-n data. The global variables and the calculations of corresponding atomic distances are now cleaner and better described in the “Build Atom” cell.
Added the horizontal/vertical z-axis orientation drop-down choice at the bottom of the gui control panel image above.
Added three new control variables: o-slots gap, epn gap and N offset. The five variable descriptions are as follows:
1. EmRadius. 2-8. The model’s proton emissions display size, or the radius of either the plane circle emission or open circular end of the cone emissions. Of course emissions go on forever, the radius is arbitrary. A larger emission setting shows how the emissions are oriented with respect to adjacent or nearby atomic particles.
2. o-slots gap. 2-3. The separation distance between adjacent orthogonal proton stacks (or proton groups).
3. epn gap. 2-4. The separation between adjacent proton groups (epn’s) within a stack.
4. N offset. 2-6. The neutron orbital radius about the direct proton pole to proton pole charge channel.
5. Cone ang (angle). 20-40 (degrees). The +/- latitudes of maximum proton charge/anticharge emissions.

Discussion. Plane emissions. Plus and minus some angle of course. I see the circular display of the proton’s “plane emissions” as a summation of both charge and anti-charge, both present in the cross-sectional proton emission field out to some radius beyond the proton’s equator. That option is of course available. I currently believe the idea of using a single emission plane to express the true size of the proton’s emission field within the simulation’s 3D model space to be too limited. Overlapped north and south torroids are better than a plane.
Cone emissions. I believe protons have +/- latitudes of maximum charge and anti-charge emissions of varying latitude width, that may be represented by two opposing cone shapes. When cone emissions are selected, both plane emissions and +/- cone emissions are included. From the side the planes might not be visible but they are there. The central dark circle shows both overlapping plane and cone emissions for the front/back aligned x-axis proton stacks. I believe the proton’s non-planar emissions may help explain proper neutron positioning and adjacent e-p-n (proton) stack slot bonding. Proper spacing between and within proton stacks or individual alphas.
Rather than subject the user entirely to my interpretations, give her controls to play with her own ideas. Easy enough to change the control widget limits as well.
Previously, I tried to code the neutron orbital position so as to avoid the proton’s +/-30 deg charge emission. Unfortunately (or not), the cones appear to suggest that neutrons are fed charge not from the direct proton pole to proton pole charge channel, but from the nearest proton emission latitude. That may be a delightful possibility (or not). In any case, the model is intended to be as accurate as possible and to stoke charge field thinking. I do not wish to corrupt the user with false model presumptions and assertions of my own. In adding controls, the user may be able to see their own charge field possibilities and less as a simulated model.
No pythreejs rendered output. Issue #381.
After a month with no problem, on 9 August, I’d lost the rendered output again. My note at the time - With the Jupyter Noteook program open in a separate window, I used my chrome browser to go to a blog site I’ve routinely visited since starting this project. The site appeared, momentarily went black - visually striking, then re-appeared normally. I went straight back to the Jupyter Notebook chrome window and saw that the threejs rendered output was gone. I tried relaunching anaconda and notebook and my notebook file – no joy. I tried relaunching chrome, anaconda notebook and my file, and saw that the rendered output was restored. Only with chrome? Relaunching chrome (and Anaconda and Notebook) is certainly easier than rebooting my computer. See if it happens again.
I’d not added any information to issue #381, yet was happy to receive a message from vidartf first thing on 10 August. The next time this issue occurs, please check your browser's console for any output (and share it here). It might be that you are hitting some browser limit related to WebGL.
An out of the blue timely Sherlock Holmes-like instruction, consistent with the facts before me. If we have a size issue now, what about doubling and tripling atoms?
I admit I’ve become a bit shy of using my browser for other things while the program is running. check your browser's console for any output (and share it here) sounds challenging.
Later, with the program running, I used another chrome window to visit that same site again. Sure enough, again saw a black flash. This time however, I tried moving my mouse to the window sliders to change the window size and somehow screwed-up and locked the screen, presumably too many commands while downloading loading a large number of images; the site also has a black background twitter feed sidebar that kicks in with a wide screen window setting. After several seconds I tried and failed to access my task manager. A minute later I ended up pressing the computer’s power switch for over 15 seconds to force it to turn off.
I tried again. The site and the program are currently both running in the background with no problem. I’ll operate as usual for a few days to see if it happens again more gracefully.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Sorry Cr6. No new progress or pythreejs output rendering disappearances to report. vidartf’s reply, “please check your browser's console for any output (and share it here)” made me check my browser’s console. Its found under “developer tools”. How embarrassing that I hadn't considered it before. The console shows Jupyter notebook messages for all the notebook projects. Before anything else, I need to study up on them and try to understand what the heck they’re telling me.
Your thoughts are always welcome.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Began at active AtomBuilder3’s cell 10, ‘Atom data’. Cleared console and ran notebook’s - Run all cells above, command. The console showed 2 verbose messages. Selected 63 Europium, and cones and then - Run all cells below - resulted in 714 messages, 707 user messages, 1 info and 713 verbose. Using the orbital camera to move the image resulted in the console output shown. Turning on a rotation generates streams of increasing numbers of messages.
More no new progress and no new pythreejs output rendering disappearances to report. Jupyter notebook is always running in the background. Trying to learn about dev tools.
“please check your browser's console for any output (and share it here)”
Jupyter notebook runs in a browser, such as chrome. The browser console is found on one of the tabs included in chrome’s built-in “developer tools” application. The easiest way (aside from using control keys) to launch the dev tools app is by right-clicking something on the browser, then clicking on ‘inspect’. The console will then usually be occupying part of the screen, it displays info status messages, alerts, warnings and errors. The console also allows live code interaction for use in site inspection, development, debugging or error correction.
Searching on both “Jupyter notebook and Developer tools” I found only one return, a gitbook. The Book of Jupyter,
https://carreau.gitbooks.io/jupyter-book/content/introtojs.html
which provides a brief overview of Developer tools, but little of any potential use that I could see. Associated tutorial(s) is alluded to but are not shown in other sections. Maybe this gitbook is a reference for said tutorial? Maybe the gitbook needs to be purchased first? Its not a reference I’d care to pursue or recommend further.
Moving along, I reviewed several “developer tools” youtube videos. As I understand it, developer tools allows one to view (or modify) the files used to create and display a web site, primarily javascript, HTML5 and CSS type files. Dev tools is intended for, maybe even absolutely essential to site developers.
The great thing about Jupyter notebook is that Jupyter notebook itself may be considered an integrated development environment (ide) which provides ‘dev tools’-like functionality in a browser automatically, such as interaction with “live” code.
Nevertheless, learning dev tools seems to offer its advantages, applicable to most any site. Jupyter notebook provides dev tools with plenty to mess with, starting with console log messages. We do have errors (not shown here) which I do not understand. I’ll need to spend more time at it.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

A failed alternative emission field model.
No code progress to speak of.
Back on 11 August, talking about proton emissions in 3D model space, I criticized circular emission planes, saying. “Overlapped north and south toroids are better than a plane.”
I attempted to replace the proton’s planar emission with some sort of toroidal emission field(s), north or south of various sizes or overlapped into single toroid's. I stand corrected, a toroidal emission is always confusing. A circular plane emission is much better than a toroid(s) emission.
Progress with respect to lost outputs.
The issue is closed.
Notebook lost the rendered output again on Monday morning, 29 August 22.
I posted a reply to vidartf, https://github.com/jupyter-widgets/pythreejs/issues/381
including a lengthy description and comment:
I wrote. Your WebGL limit guess appears to be correct. The console log includes the message: mBuilder.ipynb:1 WebGL: CONTEXT_LOST_WEBGL: loseContext: context lost.
As vidartf had directed, I also included a dev tools console log file, plus 2 more for good measure.
1. The console log file output during the loss of threejs rendered output. localhost-1661782215490.log
2. A normal initial log output. localhost-1661785463375.log
3. A normal operational log output. localhost-1661789658096.log
vidartf replied on Wednesday morning, 31 Aug 22.
Thanks for the context and the confirmation via logs. I think we can still improve our handling of context loss (https://www.khronos.org/webgl/wiki/HandlingContextLost), but in general, saving your notebook and refreshing the page should hopefully be enough.
Airman. In other words, No problemo. An occasional loss of the WebGL context (and notebook’s pythreejs rendered output) when the program is opened for many hours at a time in the background is NOT the fatal error I imagined; good to know. I thanked him and closed issue #381.
And Progress with respect to errors.
5 Initial errors.
- Code:
5 Initial errors.
main.min.js?v=7e50cb…744d71ff0834f:59830 Failed to render mimetype "application/vnd.jupyter.widget-view+json" with: Error: A Jupyter widget could not be displayed because the widget state could not be found. This could happen if the kernel storing the widget is no longer available, or if the widget state was not saved in the notebook. You may be able to create the widget by running the appropriate cells.
Last time I wrote, "We do have errors (not shown here) which I do not understand".
I should have mentioned that the dev tools console log errors received occur only during the mBuilder or AB3 notebook programs’ start, before going to cell 10 and using the run-all-above command. There’s likely some way to clear that initial error, such that the program starts with the periodic table and gui control panel displayed. In which case the user wouldn’t need to run-all-above. Maybe a future action-item. For the time being, in the dev tools videos I’ve seen, the presenter(s) tells the viewer to just clear the initial errors. After doing so, the program generates the thousands of error-less dev tool console log messages I described last week.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Looks like I'm able to render okay if "number" is hard coded and not pulled from the gui. Makes me think the gui is consuming too much memory from the initial look at it.
This may be the issue? An old K3D...they have some pretty complex scripts still rendering.
https://github.com/K3D-tools/K3D-jupyter/blob/main/README.md
You may want to ask vidartf to ensure he has the latest K3D library. BTW, I was able to render your mBuild file on my windows system with Python 3.9 with the latest Microsoft Edge browser.

Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

24 Chromium, Cr. Latest e-p-n proton stack version.
How did you hard code it? In your image, the selected tab includes a blank atom label, and the two SL tabs indicate He but I recognize Chromium when I see it. The Peg neutrons opposite the protons in slots 10 – 13 and zero neutrons in slots 14 and 15 indicate a version earlier than the current e-p-n stacks. Almost all of those e-p-n stacks currently contain single offset neutrons, orbiting the direct proton pole to proton pole charge channel. The hard coded modification was made to AB2.Cr6 wrote. Looks like I'm able to render okay if "number" is hard coded and not pulled from the gui. Makes me think the gui is consuming too much memory from the initial look at it.
The easiest code change might be to go to the “Atom data” cell, and add “number = 24” (or some other atomic number) just above, “An = elements[number-1]”. But then the tabs would indicate Cr and not He.
I don’t think gui memory is the problem. The error I posted last time includes, QUOTE. “A Jupyter widget could not be displayed because the widget state could not be found”.
https://ipywidgets.readthedocs.io/en/stable/embedding.html#save-notebook-widget-state
Here’s a site that describes widget states. If the widget state is properly saved, then just turning on the notebook file will display a rendered widget. Since learning about them yesterday, I’ve tried clearing and saving widget states without any successful saves. I keep getting broken links, lost kernals and new errors. Maybe I’m not going about it properly. The code uses several widgets: the periodic table, gui, slotLayouts, tab enclosure and rotation controls. Which widget state gets “saved” exactly?
Cr6 wrote. This may be the issue? An old K3D...they have some pretty complex scripts still rendering.
https://github.com/K3D-tools/K3D-jupyter/blob/main/README.md
You may want to ask vidartf to ensure he has the latest K3D library.
Please note that vidartf is one of the seven K3D-jupyter contributors shown on your K3D README.md link. I'm sure he's current, and then some. The "Interactive showcase gallery" link included in the README worked fine for me.
None of the AtomBuilder or mBuilder files use K3D. I recall you pointing out the three orthogonal backplanes, the K3D background grid in the past. Do you still want that? Or is there some other K3D shape you’re interested in?
Cr6 wrote. BTW, I was able to render your mBuild file on my windows system with Python 3.9 with the latest Microsoft Edge browser.
Good to know it works! Does that mean the latest Microsoft Edge browser can run Jupyter notebook files like chrome can? Or is there a python/notebook readable app like binder built-in? I gave up looking at the new Microsoft Edge browser when I saw students can pay the cheapest 50% rate at $2.99 a month.
Cr6, You’re the only one I know who’s “run” these notebook files. Please confirm you’re not having difficulties with any of them. Sure I’m making slow progress, including errors and lost outputs, I feel a bit more secure. Finally beginning to think about how I should bind two atoms together.
Pardon me for asking, I’d greatly appreciate some words, direction or critical assessment from you.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Let me do a much more thorough check on my system this weekend.
I can say it runs on Jupyter-Brave and Edge on Windows. It works fine on my Linux Mint box with Brave if the "number=XX" is hardcoded as well. Good to know about K3D and support of this library! Didn't know that.
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

I also had to make a raw call to the file with Jupyter Labs - the "Tab Crash" was the Periodic Table rendering showing "Out of Memory":

Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Mercury console log.
Thanks Cr6, I'll make this a quick update too. Hearing you’ve got “out-off memory” issues that eventually work sounds important but not debilitating. Or am I mistaken? Are you getting alerts or log messages to that affect? Any one program more than another?
I still haven’t properly saved any widget state yet, nor considered or discussed all the programs’ remaining console log warnings. They’ve put me in a bit of a funk, a state of confusion with no idea how to correct or otherwise account for them.
When spinning threejs model objects, one must assign the objects into one or more spin groups. The atom is the first spin group. For each of the up-to 19 slots to spin independently (each around their own central spin axis), each slot must be its own spin group, 19 groups. Note that every object (electron, proton and neutron) within each slot’s spin group spin in the same direction as a single object. The carousel is another spin group.
When I first began trying to code a second atom, I copied AtomBuilder2’s full set of atom, carousel and 19 SL groups. When I had that second set of spin groups available, I realized that would allow any particular proton within a slot to spin in one of two possible contra spinning directions – left or right. That enabled me to redefine an atom’s proton stack bonds based on Miles’ Diatomic Hydrogen paper. I’m happy to have arrived at the dual directional e-p-n proton stacks model although I’m sure I haven’t assigned those e-p-n stacks properly yet either.
At which time AB2 – with single directional slots, was replaced with AtomBuilder3 - dual directional e-p-n slots (mBuilder also has e-p-n slots). With respect to spin groups, the memory load of AB3 is twice that of AB2, but I haven’t noticed any “performance” difference between the two. Apparently the “out-off memory" issues do not involve spin groups. In going forward to two and three atoms, the total number of spin groups will double and triple. I’ll likely start by building simple non-spinning molecules for starters.
But first; Must concentrate on this console stuff.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
- Code:
##################################
# AtomBuilder.
# Partial transcribed outputs taken from the Console running each
# cell steps 1-22, (the guiwidgets cell twice), obtaining console
# outputs, cleaned-up/copied most of the pertinent outputs below,
# then cleared the console to run the next cell.
# Only the 1a. output is dependably constant, as well as many [Violation]s
# that seem to be assoc with mouse actions(?).
# All the other outputs including b. c. and d.are received intermittently.
##################################
AtomBuilder.
1. a. Open AtomBuilder notebook and console. Obtained outputs
labeled a. below, including: 23 mess, 21 user mess, 7 errors,
2 warnings, 13 info and 1 verbose. Cleared console.
2. Ran Atom Builder Markdown cell. Received 1 mess/verbose.
[Violation] 'click' handler took 158ms
cleared console.
3. Ran cell 'from pythreejs import ... '. Received 1 mess/verbose.
[Violation] 'click' handler took 152ms
Cleared console.
4. Ran cell 'view_width = 600 ... 'Received 2 messages/verbose.
[Violation] 'click' handler took 678ms
[Violation] Forced reflow while executing JavaScript took 230ms
Cleared console.
5. Ran cell '# Creating the periodic table'. Received 4 messages/verbose.
[Violation] 'focus' handler took 280ms
[Violation] Forced reflow while executing JavaScript took 96ms
[Violation] 'click' handler took 322ms
[Violation] Forced reflow while executing JavaScript took 60ms
Cleared console.
6. Ran cell 'Defining the GUI widgits. No console outputs.
7. Ran the Periodic table markup cell. Received 1 messages/verbose.
[Violation] 'click' handler took 176ms
Cleared console.
8. Ran cell 'grid # Display the Periodic Table'. No console outputs.
9. Ran cell 'guiwidgets'. No console outputs.
10. With the gui now active, chose an atom and ran the 'guiwidgets' cell again. Received 2 message/verbose.
No console outputs.
11. Ran Cell "This short vertical extra notebook cell ... . Received 2 more messge/verbose.
[Violation] 'click' handler took 2046ms
[Violation] Forced reflow while executing JavaScript took 783ms
Cleared console.
12. b. Ran cell 'atomicNumber = number '. Received 142 mess, 138 user mess, 142 verbose.
Reported under b. below. Cleared console.
13. Ran cell # Create Slotlayout Diagram. No console output.
14. Ran Markup cell, 'The Slot Layout diagram'. Received 1 message/verbose
[Violation] 'click' handler took 182ms
Cleared console.
15. Ran Cell 'sgrid'. Received 1 messge/verbose.
[Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
Cleared console.
16. Ran cell 'if atomsLabel.value == True:'. Received 2 messages/verbose
[Violation] 'click' handler took 186ms
[Violation] Forced reflow while executing JavaScript took 41ms
Cleared console.
17. Ran cell '# Defining the rotation ... '. Received 1 message/verbose
[Violation] Forced reflow while executing JavaScript took 42ms
Cleared console.
18. Ran markup cell, 'Carousel and Slot Rotation Controls'. No console output.
19. Ran cell 'rgrid'. Received 1 message/verbose
[Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
Cleared console.
20. c. Ran cell 15. 'renderer'. Received 9 mess, 8 user mess, 1 info and 8 verbose,
reported as c. below.
Cleared console.
20, Ran cell 19. 'rgrid'. Received 3 message/verbose.
[Violation] 'click' handler took 222ms
[Violation] Forced reflow while executing JavaScript took 53ms
[Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
Cleared console.
21. d. Ran cell '# The periodic table',the non-interactive one displayed
as a notebook tab. Received 12 mess, 1 user mess, 1 info and 11 verbose,
reported as d. below. All these outputs
22. Ran final Markup cell. No console outputs.
AtomBuilder a-d outputs:
a. Messages: 23. The 7:errors and 2:warnings and load extension:info are repeated below.
main.min.js?v=7e50cb…744d71ff0834f:69956 actions jupyter-notebook:find-and-replace does not exist, still binding it in case it will be defined later...
main.min.js?v=7e50cb…744d71ff0834f:24190 load_extensions
Arguments(7)
main.min.js?v=7e50cb…744d71ff0834f:57865 Loaded moment locale en
main.min.js?v=7e50cb…744d71ff0834f:63095 Session: kernel_created (f51e9e4e-905b-409e-80ea-802f23e22b15)
main.min.js?v=7e50cb…744d71ff0834f:62240 Starting WebSockets: ws://localhost:8888/api/kernels/411c3901-3c36-4925-8220-6cb46ee3532e
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: bqplot/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: ipycanvas/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: ipyevents/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: jupyter-datawidgets/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: jupyter-matplotlib/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: jupyter-threejs/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: jupyter-js-widgets/extension
main.min.js?v=7e50cb…744d71ff0834f:61883 Kernel: kernel_connected (411c3901-3c36-4925-8220-6cb46ee3532e)
main.min.js?v=7e50cb…744d71ff0834f:61883 Kernel: kernel_ready (411c3901-3c36-4925-8220-6cb46ee3532e)
7
main.min.js?v=7e50cb…744d71ff0834f:59830 Failed to render mimetype "application/vnd.jupyter.widget-view+json" with: Error: A Jupyter widget could not be displayed because the widget state could not be found. This could happen if the kernel storing the widget is no longer available, or if the widget state was not saved in the notebook. You may be able to create the widget by running the appropriate cells.
at extension.js:157:21
at OutputArea.<anonymous> (extension.js:168:9)
at OutputArea.append_mime_type (main.min.js?v=7e50cb…d71ff0834f:59827:43)
at OutputArea.append_display_data (main.min.js?v=7e50cb…d71ff0834f:59788:18)
at OutputArea.append_output (main.min.js?v=7e50cb…d71ff0834f:59469:18)
at OutputArea.fromJSON (main.min.js?v=7e50cb…d71ff0834f:60149:18)
at CodeCell.fromJSON (main.min.js?v=7e50cb…d71ff0834f:61382:30)
at Notebook.render_cell_output (main.min.js?v=7e50cb…d71ff0834f:67043:19)
at manager.js:131:39
at Array.forEach (<anonymous>)
DevTools failed to load source map: Could not load content for http://localhost:8888/static/notebook/js/main.min.js.map: HTTP error: status code 404, net::ERR_HTTP_RESPONSE_CODE_FAILURE
main.min.js?v=7e50cb…c744d71ff0834f:6713 [Violation] 'setTimeout' handler took 82ms
b. messages: 142
[Violation] 'focus' handler took 994ms
[Violation] Forced reflow while executing JavaScript took 366ms
[Violation] 'click' handler took 415ms
[Violation] Forced reflow while executing JavaScript took 101ms
Three.js:281 execThreeObjMethod: rotateZ(2.684218523279832)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(2.161256903808503)
Three.js:332 sending return value to server...
...
c. messages: 9.
[Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
add @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
ke @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
on @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
KeyboardManager.register_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:72326
(anonymous) @ extension.js:167
OutputArea.append_mime_type @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59827
OutputArea.append_display_data @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59788
OutputArea.append_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59469
OutputArea.handle_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59380
output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61213
Kernel._handle_output_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62975
i @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
Kernel._handle_iopub_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:63015
Kernel._finish_ws_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62794
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62785
Renderable.js:517 TV(228668): unfreeze
Renderable.js:517 TV(228668): ThreeView.acquiring...
RendererPool.js:134 RendererPool.acquiring...
three.js?v=20220913083405:179 THREE.WebGLRenderer 97
RendererPool.js:179 RendererPool.acquire(id=0)
Renderable.js:517 TV(228668): ThreeView.acquireRenderer(0)
Renderable.js:517 TV(228668): Enable controls
Renderable.js:517 TV(228668): renderScene
d. messages: 12
RendererPool.release(id=1)
Renderable.js:517 TV(914054): ThreeView WebGL context is being reclaimed: 1
Renderable.js:513 TV(914054): already frozen...
Renderable.js:517 TV(914054): Disable controls
[Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
add @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
ke @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
on @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
KeyboardManager.register_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:72326
(anonymous) @ extension.js:167
OutputArea.append_mime_type @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59827
OutputArea.append_display_data @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59788
OutputArea.append_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59469
OutputArea.handle_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59380
output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61213
Kernel._handle_output_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62975
i @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
Kernel._handle_iopub_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:63015
Kernel._finish_ws_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62794
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62785
Renderable.js:517 TV(893118): unfreeze
Renderable.js:517 TV(893118): ThreeView.acquiring...
RendererPool.js:134 RendererPool.acquiring...
RendererPool.js:179 RendererPool.acquire(id=1)
Renderable.js:517 TV(893118): ThreeView.acquireRenderer(1)
Renderable.js:517 TV(893118): Enable controls
Renderable.js:517 TV(893118): renderScene
a. User messages: 21. The user messages are the same as the messages, minus the
last two messages, DevTools failed ... and [Violation] 'setTimeout' handler took ...
b. User messages: 138. The same as message/verbose minus [Violation] Forced reflow
c. User messages: 8 Same as messages minus the first violation.
d. User messages: 11 Same as messages minus the first violation.
a. Errors: 7
Error: A Jupyter widget could not be displayed because the widget state could not be found. This could happen if the kernel storing the widget is no longer available, or if the widget state was not saved in the notebook. You may be able to create the widget by running the appropriate cells.
b. Errors: 0.
c. Errors. 0
d. Errors. 0
a. Warnings: 2.
actions jupyter-notebook:find-and-replace does not exist, still binding it in case it will be defined later...
DevTools failed to load source map: Could not load content for http://localhost:8888/static/notebook/js/main.min.js.map: HTTP error: status code 404, net::ERR_HTTP_RESPONSE_CODE_FAILURE
b. Warnings: 0.
c. Warnings: 0.
d. Warnings: 0.
a. Ifo: 13.
load_extensions Arguments(7)
Loaded moment locale en
Session: kernel_created (f51e9e4e-905b-409e-80ea-802f23e22b15)
Starting WebSockets: ws://localhost:8888/api/kernels/411c3901-3c36-4925-8220-6cb46ee3532e
Loading extension: bqplot/extension
Loading extension: ipycanvas/extension
Loading extension: ipyevents/extension
Loading extension: jupyter-datawidgets/extension
Loading extension: jupyter-matplotlib/extension
Loading extension: jupyter-threejs/extension
Loading extension: jupyter-js-widgets/extension
Kernel: kernel_connected (411c3901-3c36-4925-8220-6cb46ee3532e)
Kernel: kernel_ready (411c3901-3c36-4925-8220-6cb46ee3532e)
b. Info: 0.
c. Info: 1.
THREE.WebGLRenderer 97
c. Info: 1.
d. Info: 1
TV(914054): already frozen...
a. verbose: 1. [Violation] 'setTimeout' handler took 82ms
b. Verbose: 142. Same as Messages
c. Verbose: 8, I don't see the difference with Messages.
d. Verbose: 11, I don't see the difference with Messages.
##################################
# Partial transcribed notes taken from the Console output running
# AtomBuilder2, AtomBuilder3 and mBuilder.
# For each of these three notebooks, the outputs are obtained by
# copying then clearing the console after the following three actions:
# a. Turn on the notebook, open the console.
# b. Start at the 'Atom data' cell and run-all-above.
# c. Select atom and run-all-below.
# Additional events, like clicking on the atom data cell may also
# create an output. Not all outputs are always received.
##################################
AtomBuilder2
a. 20 mess, 18 user mess, 4 errors, 2 warnings, 13 info and 2 verbose.
b. 2 messages, 2 verbose.
c. 66 mess, 62 user mess, 1 info and 65 verbose.
The 3 sets of outputs are detailed below.
1. Two messgaes(verbose) received when I activate cell 11 - 'Atom data'
[Violation] 'mousedown' handler took 195ms
[Violation] Forced reflow while executing JavaScript took 37ms
Cleared console.
2. b. Two messgaes(verbose) received with cell run-all-above cmd.
[Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
add @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
ke @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
on @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
KeyboardManager.register_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:72326
(anonymous) @ extension.js:167
OutputArea.append_mime_type @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59827
OutputArea.append_display_data @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59788
OutputArea.append_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59469
OutputArea.handle_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59380
output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61213
Kernel._handle_output_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62975
i @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
Kernel._handle_iopub_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:63015
Kernel._finish_ws_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62794
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62785
[Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
a. Messages: 21
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:69956 actions jupyter-notebook:find-and-replace does not exist, still binding it in case it will be defined later...
MenuBar.bind_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:69956
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:24190 load_extensions Arguments(7)
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:57865 Loaded moment locale en
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:63095 Session: kernel_created (2b4ae78c-bc90-44c1-8350-14a27cce2aa7)
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62240 Starting WebSockets: ws://localhost:8888/api/kernels/7ad8fa01-4b0b-4841-8820-1bcb3afac2ce
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:24167 Loading extension: bqplot/extension
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:24167 Loading extension: ipycanvas/extension
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:24167 Loading extension: ipyevents/extension
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:24167 Loading extension: jupyter-datawidgets/extension
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:24167 Loading extension: jupyter-matplotlib/extension
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:24167 Loading extension: jupyter-threejs/extension
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:24167 Loading extension: jupyter-js-widgets/extension
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61883 Kernel: kernel_connected (7ad8fa01-4b0b-4841-8820-1bcb3afac2ce)
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61883 Kernel: kernel_ready (7ad8fa01-4b0b-4841-8820-1bcb3afac2ce)
4main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59830 Failed to render mimetype "application/vnd.jupyter.widget-view+json" with: Error: A Jupyter widget could not be displayed because the widget state could not be found. This could happen if the kernel storing the widget is no longer available, or if the widget state was not saved in the notebook. You may be able to create the widget by running the appropriate cells.
at extension.js:157:21
at OutputArea.<anonymous> (extension.js:168:9)
at OutputArea.append_mime_type (main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59827:43)
at OutputArea.append_display_data (main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59788:18)
at OutputArea.append_output (main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59469:18)
at OutputArea.fromJSON (main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:60149:18)
at CodeCell.fromJSON (main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61382:30)
at Notebook.render_cell_output (main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:67043:19)
at manager.js:131:39
at Array.forEach (<anonymous>)
OutputArea.append_mime_type @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59830
DevTools failed to load source map: Could not load content for http://localhost:8888/static/notebook/js/main.min.js.map: HTTP error: status code 404, net::ERR_HTTP_RESPONSE_CODE_FAILURE
[Violation] 'setTimeout' handler took 124ms
[Violation] Forced reflow while executing JavaScript took 48ms
b. Messages: 2
[Violation] 'mousedown' handler took 169ms
[Violation] Forced reflow while executing JavaScript took 30ms
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2 [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
add @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
ke @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
on @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
KeyboardManager.register_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:72326
(anonymous) @ extension.js:167
OutputArea.append_mime_type @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59827
OutputArea.append_display_data @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59788
OutputArea.append_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59469
OutputArea.handle_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59380
output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61213
Kernel._handle_output_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62975
i @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
Kernel._handle_iopub_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:63015
Kernel._finish_ws_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62794
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62785
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2 [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive
c. Messages: 66.
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2 [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
add @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
ke @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
on @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
KeyboardManager.register_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:72326
(anonymous) @ extension.js:167
OutputArea.append_mime_type @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59827
OutputArea.append_display_data @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59788
OutputArea.append_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59469
OutputArea.handle_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59380
output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61213
Kernel._handle_output_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62975
i @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
Kernel._handle_iopub_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:63015
Kernel._finish_ws_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62794
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62785
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2 [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
add @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
ke @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
on @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
KeyboardManager.register_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:72326
append_html @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59853
OutputArea.append_mime_type @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59827
OutputArea.append_execute_result @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59617
OutputArea.append_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59449
OutputArea.handle_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59380
output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61213
Kernel._handle_output_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62975
i @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
Kernel._handle_iopub_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:63015
Kernel._finish_ws_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62794
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62785
Three.js:281 execThreeObjMethod: rotateZ(3.6601345488551904)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(3.9606423177172725)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(3.541708138013911)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateX(3.144605511029693)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(1.1291024138801349)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(3.996000721381145)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(1.6823098226098958)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(4.448500898796295)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateX(3.144605511029693)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(5.4799944322193905)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateX(-1.5723027555148466)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(3.7096670904510565)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(3.483488269522218)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(2.7623303612139223)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateX(3.144605511029693)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(5.0784759369562815)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateX(-1.5723027555148466)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(3.710645407583361)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(5.357442920890825)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(1.0079073307971038)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateX(3.144605511029693)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(5.793526966252417)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(5.383785258131928)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(5.203488784066734)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(2.5614657683906366)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateX(3.144605511029693)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(2.6914257693249666)
Three.js:332 sending return value to server...
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2 [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
add @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
ke @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
on @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
KeyboardManager.register_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:72326
(anonymous) @ extension.js:167
OutputArea.append_mime_type @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59827
OutputArea.append_display_data @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59788
OutputArea.append_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59469
OutputArea.handle_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59380
output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61213
Kernel._handle_output_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62975
i @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
Kernel._handle_iopub_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:63015
Kernel._finish_ws_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62794
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62785
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2 [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event. Consider using MutationObserver to make the page more responsive.
add @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
ke @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
on @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
KeyboardManager.register_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:72326
(anonymous) @ extension.js:167
OutputArea.append_mime_type @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59827
OutputArea.append_display_data @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59788
OutputArea.append_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59469
OutputArea.handle_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59380
output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61213
Kernel._handle_output_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62975
i @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
Kernel._handle_iopub_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:63015
Kernel._finish_ws_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62794
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62785
Renderable.js:517 TV(973851): unfreeze
Renderable.js:517 TV(973851): ThreeView.acquiring...
RendererPool.js:134 RendererPool.acquiring...
three.js?v=20220915103453:179 THREE.WebGLRenderer 97
RendererPool.js:179 RendererPool.acquire(id=0)
Renderable.js:517 TV(973851): ThreeView.acquireRenderer(0)
Renderable.js:517 TV(973851): Enable controls
Renderable.js:517 TV(973851): renderScene
a. User messages: 18.
b. User messages: 0.
c. User messages: 62.
b. Errors: 0
c. Errors: 0
a. Warnings: 2
main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:69956 actions jupyter-notebook:find-and-replace does not exist, still binding it in case it will be defined later...
MenuBar.bind_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:69956
DevTools failed to load source map: Could not load content for http://localhost:8888/static/notebook/js/main.min.js.map: HTTP error: status code 404, net::ERR_HTTP_RESPONSE_CODE_FAILURE
b. Warnings: 0
c. Warnings: 0
a. Info: 13
main.min.js?v=7e50cb…744d71ff0834f:24190 load_extensions
Arguments(7)
main.min.js?v=7e50cb…744d71ff0834f:57865 Loaded moment locale en
main.min.js?v=7e50cb…744d71ff0834f:63095 Session: kernel_created (2b4ae78c-bc90-44c1-8350-14a27cce2aa7)
main.min.js?v=7e50cb…744d71ff0834f:62240 Starting WebSockets: ws://localhost:8888/api/kernels/7ad8fa01-4b0b-4841-8820-1bcb3afac2ce
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: bqplot/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: ipycanvas/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: ipyevents/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: jupyter-datawidgets/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: jupyter-matplotlib/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: jupyter-threejs/extension
main.min.js?v=7e50cb…744d71ff0834f:24167 Loading extension: jupyter-js-widgets/extension
main.min.js?v=7e50cb…744d71ff0834f:61883 Kernel: kernel_connected (7ad8fa01-4b0b-4841-8820-1bcb3afac2ce)
main.min.js?v=7e50cb…744d71ff0834f:61883 Kernel: kernel_ready (7ad8fa01-4b0b-4841-8820-1bcb3afac2ce)
b. Info: 0.
c. Info: 1.
THREE.WebGLRenderer 97
a. verbose: 2
[Violation] 'setTimeout' handler took 124ms
[Violation] Forced reflow while executing JavaScript took 48ms
b. Verbose: 2. see messages.
c. Verbose: 82. see messsages.
##################################
.Cr6 wrote. Unfortunately, we may have to go to a "Graph" database with this rather than a simple 2D matrix with a table. It is a whole re-engineering to a degree but it should allow more configurations with 3-D work.
Cr6, I did some database reviews. At present, the data is in a csv file and each atom is assembled according to the build atom cell. I suppose AtomBuilder3 might be described as a Document (key:value) database, each different atom (key), taken from a table has a unique model output (value). A Graph database is described as being most appropriate for identifying relationships between many-to-many objects. A molecule selected by picking and bonding atoms from the many available atoms and bonding locations should qualify. I believe you’re correct, a graph database is the way to go.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
A couple of graph databases I'm looking at are Neo4j, OrientDB, HugeGraph and SurrealDB. Just looking for something small (free) and effective. Also looking to see what can be leveraged with Python-Jupyter that you have already created.
There are a few of them out there:
https://www.libhunt.com/topic/graph-database
https://github.com/apache/incubator-hugegraph
https://github.com/orientechnologies/orientdb
https://memgraph.com/blog/memgraph-with-python-and-jupyter-notebooks
More examples:
https://solutionsreview.com/data-management/the-best-graph-databases/
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
- Code:
#####################################################
# Summary Partial transcribed Console log outputs for:
# AtomBuilder, AtomBuilder2, AtomBuilder3 and mBuilder.
###########################################
# Index: At the document end. Eight specific recurring
# sets of outputs of multiple lines are replaced with
# a single line index title, preceeded with a #, such as
# '# [Violation] Added ... '. The index shows the full
# set of outputs and locations where the output occurs.
# Without the index this document would double or triple
# in size, and overly complicate this summary document.
#####################################################
# AtomBuilder.
# Partial transcribed outputs taken from the Console
# running each cell, 1-23, (the guiwidgets cell is run
# twice). Resulting in theconsole outputs. Cleaned-up/
# copied most of the pertinent outputs below, then
# cleared the console to run the next cell.
# Only the 1a. output is dependably constant, as well
# as several [Violation]s that seem to be associated
# with mouse click actions. All the other outputs
# including b. c. and d.are received intermittently.
#####################################################
AtomBuilder.
1. a. Open AtomBuilder notebook and console. Obtained outputs
labeled a. below, including: 23 mess, 21 user mess, 7 errors,
2 warnings, 13 info and 1 verbose. Cleared console.
2. Ran Atom Builder Markdown cell. Received 1 mess/verbose.
[Violation] 'click' handler took 158ms
cleared console.
3. Ran cell 'from pythreejs import ... '. Received 1 mess/verbose.
[Violation] 'click' handler took 152ms
Cleared console.
4. Ran cell 'view_width = 600 ... 'Received 2 messages/verbose.
[Violation] 'click' handler took 678ms
[Violation] Forced reflow while executing JavaScript took 230ms
This number will grow with mouse clicks(?). Cleared console.
5. Ran cell '# Creating the periodic table'. Received 4 messages/verbose.
[Violation] 'mousedown' handler took 152ms
[Violation] 'focus' handler took 280ms
[Violation] Forced reflow while executing JavaScript took 96ms
[Violation] 'click' handler took 322ms
Cleared console.
6. Ran cell 'Defining the GUI widgits. No console outputs.
7. Ran the Periodic table markup cell. Received 1 messages/verbose.
[Violation] 'click' handler took 176ms
Cleared console.
8. Ran cell 'grid # Display the Periodic Table'. Received 2 messages/verbose.
[Violation] 'click' handler took 184ms
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
Cleared console.
9. Ran cell 'guiwidgets'. No console outputs.Received 2 messages/verbose.
[Violation] 'click' handler took 178ms
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
Cleared console.
10. With the gui now active, chose an atom and ran the 'guiwidgets' cell again.
Received 2 messages/verbose:
[Violation] 'click' handler took 2376ms
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
Cleared console.
11. Ran Cell "This short vertical extra notebook cell ... .
[Violation] 'click' handler took 2260ms
[Violation] Forced reflow while executing JavaScript took 955ms
Cleared console.
12. b. Ran cell 'atomicNumber = number '. Received 36 mess, 34 user mess, 36 verbose.
Reported under b. below. Cleared console.
13. Ran cell # Create Slotlayout Diagram. No console output.
14. Ran Markup cell, 'The Slot Layout diagram'. Received 1 message/verbose
[Violation] 'click' handler took 182ms
Cleared console.
15. Ran Cell 'sgrid'. Received 1 messge/verbose.
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
Cleared console.
16. Ran cell 'if atomsLabel.value == True:'. Received 2 messages/verbose
[Violation] 'click' handler took 186ms
[Violation] Forced reflow while executing JavaScript took 41ms
Cleared console.
17. Ran cell '# Defining the rotation ... '. Received 1 message/verbose
[Violation] Forced reflow while executing JavaScript took 42ms
Cleared console.
18. Ran markup cell, 'Carousel and Slot Rotation Controls'. No console output.
19. Ran cell 'rgrid'. Received 1 message/verbose
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
Cleared console.
20. c. Ran cell 15. 'renderer'. Received 9 mess, 8 user mess, 1 info and 8 verbose,
reported as c. below.
Cleared console.
21, Ran cell 19. 'rgrid'. Received 3 message/verbose.
[Violation] 'click' handler took 222ms
[Violation] Forced reflow while executing JavaScript took 53ms
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
Cleared console.
22. d. Ran cell '# The periodic table', the non-interactive one displayed
as a notebook tab. Received 12 mess, 11 user mess, 1 info and 11 verbose,
reported as d. below.
23. Ran final Markup cell. No console outputs.
AtomBuilder (AB) a-d outputs:
a. 23 Messages: 7:errors, 2:warnings, 1:# load extension(13).
# Warning. actions jupyter-notebook:find-and-replace does not exist
# load_extensions(13)
# 7 Error. Failed to render.
# DevTools failed to load source map:
[Violation] 'setTimeout' handler took 82ms
b. 36 messages: # The total number will depend on the atom selected.
[Violation] 'click' handler took 325ms
[Violation] Forced reflow while executing JavaScript took 85ms[Violation] 'focus' handler took 197ms
# Three.js execThreeObjMethod ... sending (34)
c. 9 messages:
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
8 # Renderableble. ... renderScene (8)
THREE.WebGLRenderer 97
d. 12 messages:
4 # RendererPool (4)
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# Renderableble. ... renderScene (8)
a. 21 User messages:
# Warning. actions jupyter-notebook:find-and-replace does not exist
# load_extensions(13)
7 # Error. Failed to render.
b. 34 User messages: # Three.js execThreeObjMethod ... sending (34)
c. 8 User messages: # Renderableble. ... renderScene (8). Same as messages minus the first violation.
d. 11 User messages:
4 # RendererPool (4)
# Renderableble. ... renderScene (8)
a. 7 Errors: 7 # Error. Failed to render.
b. Errors: 0
c. Errors. 0
d. Errors. 0
a. 2 Warnings:
# Warning. actions jupyter-notebook:find-and-replace does not exist
# DevTools failed to load source map: Could not load contentb. Warnings: 0.
b. Warnings: 0.
c. Warnings: 0.
d. Warnings: 0.
a. 13 Info: # load_extensions(13)
b. Info: 0.
c. Info: 1. THREE.WebGLRenderer 97
d. Info: 1. TV(914054): already frozen...
a. 1 Verbose: [Violation] 'setTimeout' handler took 82ms
b. 36 Verbose: Same as b. Messages
c. 8 Verbose: I don't see the difference with Messages.
d. 11 Verbose: I don't see the difference with Messages.
##################################
# Partial transcribed notes taken from the Console output running
# AtomBuilder2, AtomBuilder3 and mBuilder.
# For each of these three notebooks, the outputs are obtained by
# copying then clearing the console after the following three actions:
# a. Turn on the notebook, open the console.
# b. Start at the 'Atom data' cell and run-all-above.
# c. Select atom and run-all-below.
# Additional events, like clicking on the atom data cell may also
# create an output. Not all outputs are always received.
##################################
AtomBuilder2
a. 27 mess, 18 user mess, 4 errors, 2 warnings, 13 info and 2 verbose.
b. 2 messages, 2 verbose.
c. 78 mess, 73 user mess, 1 info and 77 verbose.
The 3 sets of outputs are detailed below.
1. Two messgaes(verbose) received when I activate cell 11 - 'Atom data'
[Violation] 'mousedown' handler took 195ms
[Violation] Forced reflow while executing JavaScript took 37ms
Cleared console.
a. 27 Messages:
# Warning. actions jupyter-notebook:find-and-replace does not exist
13 # load_extensions(13)
4 # Error. Failed to render.
# DevTools failed to load source map: Could not load content
6 [Violation] 'setTimeout' handler took <N>ms
[Violation] Forced reflow while executing JavaScript took 48ms
b. 2 Messages/verbose 2:
2 # [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
c. 78 Messages:
# RendererPool (4)
[Violation] 'click' handler took 186ms
2 # [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
2 # [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# Three.js execThreeObjMethod: ... sending
2 # [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
2 # [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# Renderableble. ... renderScene (8)
a. 18 User messages:
# Warning. actions jupyter-notebook:find-and-replace does not exist
13 # load_extensions(13)
4 # Error. Failed to render.
b. User messages: 0.
c. 73 User messages:
# RendererPool (4)
# Three.js execThreeObjMethod: ... sending
# Renderableble. ... renderScene (8)
a. Errors: 4 # Error. Failed to render.
b. Errors: 0
c. Errors: 0
a. Warnings: 2
# Warning. actions jupyter-notebook:find-and-replace does not exist.
# DevTools failed to load source map: Could not load contentb. Warnings: 0
b. Warnings: 0
c. Warnings: 0
a. 13 Info: 13 # load_extensions(13)
b. Info: 0.
c. Info: 1. # THREE.WebGLRenderer 97
a. 8 verbose:
6 [Violation] 'setTimeout' handler took <N>ms
[Violation] Forced reflow while executing JavaScript took 48ms
[Violation] Forced reflow while executing JavaScript took 39ms
b. 2 Verbose: see messages.
c. 77 Verbose: see messsages.
##################################
AtomBuilder3
a. 21 mess, 19 user mess, 5 errors, 2 warnings, 13 info and 1 verbose.
b. 2 messages, 2 verbose.
c. 73 mess, 60 user mess, 1 info and 72 verbose.
The 3 sets of outputs are detailed below.
1. One messgaes(verbose) received when I activate cell 11 - 'Atom data'. The same info data as in AtomBuilder2 above.
[Violation] 'mousedown' handler took 195ms
[Violation] Forced reflow while executing JavaScript took 37ms.
Clear console hit run-all-above.
a. Messages: 21
# Warning. actions jupyter-notebook:find-and-replace does not exist
# load_extensions(13)
5 # Error. Failed to render.
# DevTools failed to load source map: Could not load content
[Violation] 'setTimeout' handler took 111ms
[Violation] Forced reflow while executing JavaScript took 37msc.
b. Messages: 3.
[Violation] 'setTimeout' handler took 57ms
[Violation] 'mousedown' handler took 167ms
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
c. 73 Messages:
[Violation] 'setTimeout' handler took 57ms
[Violation] 'click' handler took 205ms
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# Three.js execThreeObjMethod: ... sending
3 # [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# Renderableble. ... renderScene (8)
6 [Violation] 'setTimeout' handler took <N>ms
[Violation] Forced reflow while executing JavaScript took 32ms
[Violation] Forced reflow while executing JavaScript took 32ms
a. 19 User messages:
# Warning. actions jupyter-notebook.
# load_extensions(13).
5# Error. Failed to render.
b. User messages: 0
c. 60 User messages:
# Three.js execThreeObjMethod: ... sending
# Renderableble. ... renderScene (8)
THREE.WebGLRenderer 97
a. Errors: 5. # Error. Failed to render.
See index.The same type widget errors as in AtomBuilder2 above.-
b. Errors: 0.
c. Errors: 0.
a. 2 Warnings:
# Warning. actions jupyter-notebook:find-and-replace does not exist
# DevTools failed to load source map: Could not load content
b. Warnings: 0.
c. Warnings: 0.
a. 13 Info: 13. # load_extensions(13)
b. Info: 0.
c. 1 Info: THREE.WebGLRenderer 97
a. 1 verbose: [Violation] 'setTimeout' handler took 127ms
b. 3 Verbose:
[Violation] 'setTimeout' handler took 57ms
[Violation] 'mousedown' handler took 167ms
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
c. Verbose: 72. See messages minus one(?).
##################################
mBuilder.
a. 24 mess, 19 user mess, 5 errors, 2 warnings, 13 info and 4 verbose.
b. 2 messages, 2 verbose.
c. 125 mess, 117 user mess, 1 info and 124 verbose.
The 3 sets of outputs are detailed below.
1. Four messgaes(verbose) received when I activate cell 11 - 'Atom data'. The same info data as in AtomBuilder2 above.
[Violation] Handling of 'mousewheel' input event was delayed for 112 ms due to main thread being busy. Consider marking event handler as 'passive' to make the page more responsive.
[Violation] 'mousedown' handler took 235ms
[Violation] 'mousedown' handler took 284ms
[Violation] Forced reflow while executing JavaScript took 81ms
a. 24 Messages:
# Warning. actions jupyter-notebook:find-and-replace does not exist
# load_extensions(13)
5 # Error. Failed to render.
# DevTools failed to load source map: Could not load content
[Violation] 'setTimeout' handler took 137ms
[Violation] 'setTimeout' handler took 71ms
b. 2 Messages:
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
c. 125 Messages:
# RendererPool (4)
[Violation] 'click' handler took 205ms
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# Three.js execThreeObjMethod: ... sending
3 # [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# Renderableble. ... renderScene (8)
[Violation] 'setTimeout' handler took 79ms
[Violation] 'setTimeout' handler took 86ms
[Violation] 'setTimeout' handler took 89ms
a. 19 User messages:
# Warning. actions jupyter-notebook:find-and-replace does not exist
# load_extensions(13)
5 # Error. Failed to render.
b. User messages: 0.
c. 117 User messages:
# RendererPool.js
# Three.js execThreeObjMethod: ... sending
a. 5 Errors: 5 # Error. Failed to render.
b. Errors: 0
c. Errors: 0
a. 2 Warnings:
# Warning. actions jupyter-notebook:find-and-replace does not exist
# DevTools failed to load source map: Could not load content
b. Warnings: 0.
c. Warnings: 0
a. 13 Info: # load_extensions(13)
b. Info: 0.
c. 1 Info: Renderable.js:513 TV(234052): already frozen...
4 [Violation] 'setTimeout' handler took 142ms # of different durations
a. 124 verbose:
# RendererPool (4)
[Violation] 'click' handler took 205ms
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# Three.js execThreeObjMethod: ... sending
3 # [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# Renderableble. ... renderScene (8)
[Violation] 'setTimeout' handler took 79ms
[Violation] 'setTimeout' handler took 86ms
[Violation] 'setTimeout' handler took 89ms
b. 2 verbose:
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
###########################################
# Index:
# DevTools failed to load source map: Could not load content for http://localhost:8888/static/notebook/js/main.min.js.map: HTTP error: status code 404, net::ERR_HTTP_RESPONSE_CODE_FAILURE
# (AB)a.msgs,warnings, (AB2)a.msgs,warnings, (AB3) a.msgs,warnings, (mB) a.msgs,warnings
# Error. Failed to render.
# (AB)a.msgs,usermsgs,err;(AB2)a.msgs,usermsgs,err;(AB3)a.msgs,usermsgs,err;(mB)a.msgs,usermsgs,err;
Failed to render mimetype "application/vnd.jupyter.widget-view+json" with:
Error: A Jupyter widget could not be displayed because the widget state
could not be found. This could happen if the kernel storing the widget
is no longer available, or if the widget state was not saved in the
notebook. You may be able to create the widget by running the appropriate cells.
at extension.js:157:21
at OutputArea.<anonymous> (extension.js:168:9)
at OutputArea.append_mime_type (main.min.js?v=7e50cb…d71ff0834f:59827:43)
at OutputArea.append_display_data (main.min.js?v=7e50cb…d71ff0834f:59788:18)
at OutputArea.append_output (main.min.js?v=7e50cb…d71ff0834f:59469:18)
at OutputArea.fromJSON (main.min.js?v=7e50cb…d71ff0834f:60149:18)
at CodeCell.fromJSON (main.min.js?v=7e50cb…d71ff0834f:61382:30)
at Notebook.render_cell_output (main.min.js?v=7e50cb…d71ff0834f:67043:19)
at manager.js:131:39
at Array.forEach (<anonymous>)
# load_extensions(13). # A set of 13 'load_extensions ...' console outputs.
# (AB)a.msgs,usermsgs,info;(AB2)a.msgs,usermsgs,info;(AB3)a.msgs,usermsgs,info;(mBa.msgs,usermsgs,info
load_extensions > Arguments(7)
Loaded moment locale en main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:57865
Session: kernel_created (238889e7-6c84-4eea-802e-e57f4bbb74dc)
Starting WebSockets: ws://localhost:8888/api/kernels/c49d8e68-26b6-4ecf-853a-79a3a5265552
Loading extension: bqplot/extension
Loading extension: ipycanvas/extension
Loading extension: ipyevents/extension
Loading extension: jupyter-datawidgets/extension
Loading extension: jupyter-matplotlib/extension
Loading extension: jupyter-threejs/extension
Loading extension: jupyter-js-widgets/extension
Kernel: kernel_connected (c49d8e68-26b6-4ecf-853a-79a3a5265552)
Kernel: kernel_ready (c49d8e68-26b6-4ecf-853a-79a3a5265552)
# [Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
# 25+ (AB)8.9.10.15.19.23.a.c.d. msgs/verbose;(AB2)b.msgs/verbose;(AB3)b.c.msgs/a.b.verbose;(mB)b.c.msgs/verbose;
[Violation] Added synchronous DOM mutation listener to a 'DOMNodeInserted' event.
add @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
each @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
ke @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
on @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
KeyboardManager.register_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:72326
(anonymous) @ extension.js:167
OutputArea.append_mime_type @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59827
OutputArea.append_display_data @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59788
OutputArea.append_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59469
OutputArea.handle_output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:59380
output @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:61213
Kernel._handle_output_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62975
i @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:2
Kernel._handle_iopub_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:63015
Kernel._finish_ws_message @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62794
(anonymous) @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:62785
# Warning. actions jupyter-notebook: find-and-replace does not exist, still binding it in case it will be defined later...
# (mB)a. msgs/warnings
actions jupyter-notebook:find-and-replace does not exist, still binding it in case it will be defined later...
MenuBar.bind_events @ main.min.js?v=7e50cbb06bc00a057be483279bc938810b467e6c9dd9b5d91eb4b06b75704564224322da095d638da40ccb4cbe79a45d46de8eb77ac598b5c1c744d71ff0834f:69956
# Renderableble. ... renderScene (8)
# (AB)c.d.msgs,usermsgs;(AB2)c.d.msgs,usermsgs;(AB3)c.d.msgs,usermsgs;(mB)a.verbose;c.msgs;
Renderableble.js:517 TV(473661): unfreeze
Renderable.js:517 TV(473661): ThreeView.acquiring...
RendererPool.js:134 RendererPool.acquiring...
three.js?v=20220918074355:179 THREE.WebGLRenderer 97#############Not always there
RendererPool.js:179 RendererPool.acquire(id=0)
Renderable.js:517 TV(473661): ThreeView.acquireRenderer(0)
Renderable.js:517 TV(473661): Enable controls
Renderable.js:517 TV(473661): renderScene
# RendererPool (4)
# (AB)d.msgs,usermsgs;(AB2)c.msgs,usermsgs;(mB)c.msgs,usermsgs,a.verbose;
Renderable.js:186 RendererPool.js:186 RendererPool.release(id=0)
Renderable.js:517 TV(792621): ThreeView WebGL context is being reclaimed: 0
Renderable.js:513 TV(792621): already frozen...
Renderable.js:517 TV(792621): Disable controls
# Three.js execThreeObjMethod: ... sending
# (AB)b.msgs,usermsgs;(AB2)c.msgs,usermsgs;(AB3)c.msgs,usermsgs;(mB)c.msgs,usermsgs,a.verbose;
Three.js:281 execThreeObjMethod: rotateZ(3.968100637147985)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(1.6495447725964547)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(4.418584782973118)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateX(3.144605511029693)
Three.js:332 sending return value to server...
Three.js:281 execThreeObjMethod: rotateZ(5.643491790380974)
Three.js:332 sending return value to server...
Please recall I’ve stated that the dataframe’s slotlayout diagrams are not simple 2D matrices in a 90 element table. Given the nature of the data along with x, y and z coordinate information, our ‘Relational’ database is three dimensional data. I’ve agreed to a “Graph” database, but that doesn’t mean I know how it should or would work.Cr6 wrote. Unfortunately, we may have to go to a "Graph" database with this rather than a simple 2D matrix with a table.
How so? How would a graph database be useful in modeling atoms or molecules, or be used at all in this project? I agreed to a 'Graph' database without a proper understanding of what what a graph database is. It appears to me Graph databases create graph diagrams intended to display any and all simple or complex relationships between node objects, yet graph diagrams do not recreate real 3D structures. Unless the ‘Graph’ database includes its own 3D renderer, on a notebook, the 3D models will still need to be pythreejs renderings.It is a whole re-engineering to a degree but it should allow more configurations with 3-D work.
From a graph perspective (?), I suppose the lowest level nodes should be individual charge particles. Electrons, neutrons and protons.
Nodes: e, p, n.
Each individual particle needs a spin property as well as charge input and output ‘edges’ connecting that node to and from the space around the particle. Are 3D graphs possible, is the basic ‘charge particle recycling charge’ diagram a valid graph diagram? If not, It should be.
Together, the three e, p, and n nodes form a proton group. With edges between e, p, n particles: e-P, P-e, n-P, P-n, following the same AB3 T,B,R,L rules currently in place.
I suppose the next higher level node should be the slot node, comprised of 1 to 6 proton groups again with internal T,B,R,L edges.
The next higher node might be the atomic node, consisting of up to 19 slot nodes in the atom's SL configuration.
That’s pretty much how I see nodes and edges might be be organized, pretty much the same as it is now. A question of how the build atom cell is organized.
How might you apply a graph database? Do you want to add atomic or molecular relationships?
If so, what about Cytoscape?
“Cytoscape is an open source bioinformatics software platform for visualizing molecular interaction networks
and integrating with gene expression profiles and other state data.” https://en.wikipedia.org/wiki/Cytoscape
Cytoscape doesn’t appear among the 12 best considered in your solutionsreview link.
https://solutionsreview.com/data-management/the-best-graph-databases/
The 12 Best Graph Databases to Consider for 2022
Posted on December 22, 2021 by Timothy King in Best Practices
Nor is cytoscape listed among the top 23 at graph-database open-source projects at libhunt.com
https://www.libhunt.com/topic/graph-database
Doing a search at libhunt
https://www.libhunt.com/search?query=cytoscape
the top return is
Cytoscape.js.
Graph theory (network) library for visualisation and analysis.
Then there’s Ipycytoscape.
cytoscape/ipycytoscape
A widget enabling interactive graph visualization with cytoscape.js in JupyterLab and the Jupyter notebook.
https://github.com/cytoscape/ipycytoscape
Ipycytscape is a github project that may not work, but It’s free, and uses Pandas dataframes and json files. Well, I’d a chance to update the project’s original ‘untouched’ json files.
I suppose ipycytoscape is not a database, it just creates network ‘visualizations’ that look like they were created by a graph database. As such I don’t believe one can properly ‘query’ data with ipycytoscape, but I don’t believe we need queries.
On the other hand cytoscape was designed with things like molecular interaction networks in mind, if not for recreating actual 3D models then at least for demonstrating atomic or molecular graph relationships.
Thanks for giving my imagination a good kick but I still don’t see how a graph database makes building molecules any easier. I’d be happy to try and add functionality to the existing code. What am I missing?
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
I know this is still just ideas this point about a Graph Database representation.
Good points:
LTAM wrote:From a graph perspective (?), I suppose the lowest level nodes should be individual charge particles. Electrons, neutrons and protons.
Nodes: e, p, n.
Each individual particle needs a spin property as well as charge input and output ‘edges’ connecting that node to and from the space around the particle. Are 3D graphs possible, is the basic ‘charge particle recycling charge’ diagram a valid graph diagram? If not, It should be.
Together, the three e, p, and n nodes form a proton group. With edges between e, p, n particles: e-P, P-e, n-P, P-n, following the same AB3 T,B,R,L rules currently in place.
I suppose the next higher level node should be the slot node, comprised of 1 to 6 proton groups again with internal T,B,R,L edges.
The next higher node might be the atomic node, consisting of up to 19 slot nodes in the atom's SL configuration.
That’s pretty much how I see nodes and edges might be be organized, pretty much the same as it is now. A question of how the build atom cell is organized.
Frankly this needs to be represented as Vertices in a GraphDB?
The only thing that got me really thinking about this is that when it comes to "bonding" with the charge field -- essentially creating complex molecules, Graph databases may allow for easier algos with bonds. It may be trickier to do this with a 2-D table. Not that it couldn't be done in 2-D but the Graph database may allow for more coverage of calcs, especially since we are not on super powerful systems. The question is if I throw a CO2 molecule at the algos in terms of Mathis' C.F. what would I need to calc with in-line D3.js versus a query against a graph database that could return a 2-D grid that could be represented with your cool (and formerly Nevyn's cool) rendering engine with D3.js? I'm thinking of ...just let the database calc the C.F. and then with the 2-D grid returned have it interactive? These are just thoughts at this point. Your mention of Cytoscape is a definite contender for something to build with: "Load and save previously-constructed interaction networks in GML format (Graph Modelling Language)". Frankly I didn't know there was a GML until I looked up your above post. This stuff, Graph DBs, can get trickier than it often needs to be with a lot of debugging required. I just think if we go to converting traditional Molecular Bonding to Mathis' grid of bonding...we may need to look at Graph DBs (javascript maybe initially)...only because they can allow more expression of "Features" related to making bonds. Initially I was thinking we could make a molecular converter for Traditional molecular notation to Graph-Mathis styles? I know these ideas are like walking on light ice across a frozen lake but...we might make the other side with it.
A lot of the graphics with these Graph DBs is just showing Feature connections between circles...not 3-D representations of actual interactive "bonds". I've played a bit with TinkerPop and thought it might be cool to make a GraphDB for Mathis' C.F. with it -- like a converter system.
It seems like the guys that are doing Gene modeling need something similar with feature rich bonding, that can take a lot of conditional requirements, hence your mention of Cytoscape and ipycytoscape. It is definitely a massive programming effort to go this direction. Question essentially is where to place the bonding logic, in the app interface in-line, or in a query engine...and still keep it fast and true?
Just thought that GraphDBs could allow more representation of "AB3 T,B,R,L rules" between atoms essentially allowing for a GraphDB to produce "can these 2+ atoms bond?" early. We are talking "nodes and edges" with a GraphDB like with Cytoscape-- "Easily navigate large networks (100,000+ nodes and edges) by an efficient rendering engine." This OrientDB got me thinking in this direction: https://github.com/orientechnologies/orientdb https://www.tutorialspoint.com/orientdb/orientdb_python_interface.htm
They have an "emissions" jupyter notebooks: https://github.com/dritter-sap/orientdb-jupyter
https://github.com/IBM/graph-db-insights/blob/master/README.md (Hate to present extra over head with GraphDBs they are mind-game trying to get them functional for an app from what I've seen.)
https://www.kelvinlawrence.net/book/PracticalGremlin.html#whygraph
Shows some of the complexity using GraphDBs:
https://docs.janusgraph.org/schema/
Vid on OrientDB:
https://www.youtube.com/watch?v=kpLqfFGubKM
Vid on TigerGraph:
https://colab.research.google.com/drive/1fJpcv-q0NLfHj3X1k6Lbwddp8gVVcfES#scrollTo=kCaZh0DO3HNP
https://www.youtube.com/watch?v=NtNW2e8MfCQ
https://www.youtube.com/watch?v=2BcC3C-qfX4 <-- pyGraph demo
https://www.techtarget.com/searchbusinessanalytics/news/252520646/TigerGraph-unveils-new-tool-for-machine-learning-modeling
With a GraphDB it might be easier to say "Why do these three atoms bond?" but with typical def function logic it gets unwieldy to account for edge details like with:
" the three e, p, and n nodes form a proton group. With edges between e, p, n particles: e-P, P-e, n-P, P-n, following the same AB3 T,B,R,L rules currently in place."
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Some software reviewed:
Feel free to correct me. I lost my Neo4J notes somewhere.
TigerGraph Machine Learning Workbench looks good until - the free developer version may be “applied for”. Or one free service provided tigergraph scheme/solution can exist at any given time. https://info.tigergraph.com/ml-workbench-download
OrientDB – Requires an IBM Watson Studio cloud service provider.
https://github.com/orientechnologies/orientdb
Get insights from OrientDB database using PyOrient through IBM Data Science Experience (DSX)
D3.js. https://d3-graph-gallery.com/ D3.js is a javascript library that lets you build sexy 2D charts (or graphs) of data representational networks. The closest D3 gets to 3D appears to be the ridgeline type graph. https://d3-graph-gallery.com/ridgeline.html
TinkerPop. I have yet to learn a query language, SQL or NoSQL. The Gremlin graph traversal language seems simple enough. Is TinkerPoP a graphDB?
NetworkX. Next I considered the problem in terns of converting a pandas dataframe into a graphDB. It appears easy to do so with NetworkX. The graphs can be viewed with MatPlotLib, or made sexier with D3.js or GraphViz. Too good to be true?
Airman wrote:From a graph perspective (?), I suppose the lowest level nodes should be individual charge particles. Electrons, neutrons and protons...
Cr6 wrote. Frankly this needs to be represented as Vertices in a GraphDB?
Airman. I enjoyed the Vid on OrientDB. Liugi Dell’Aquila explains a graphDB well. Both he and the TinkerPop Gremlin referred to Graph Theory 101, Graph = (V,E), vertices and edges. TigerGraph describes vertexes as nodes. Vertices and nodes mean the same to me. In my e p n node description I was also assuming – perhaps incorrectly, that a graphDB would allow parent/child nested objects for expanding or collapsing graphs – something not easily done in table data.
For the record, I don’t believe we have system memory problems, but I can make some. If I had my charge-field druthers, the code would be very memory intensive. The smallest node would be a photon, with properties: position, radius, mass, velocity, direction, spin direction, spin velocity, etc. Each proton would consist of the equivalent of 6 billion recycling photons; but I don’t have a simple understanding of charge recycling within e, p, or n charge particles; imagining cyclonic motions of photons within nested spin doublings constantly recycling charge through the charged particle. I’m afraid I’ll stick to billiard ball spherical atomic particles for the time being. If I do manage to include photons, there could be plenty on the screen, but they can only be displayed outside (passing by, emerging from, disappearing into, or bouncing off) e, p, or n charge particle spherical surfaces.
Cr6 wrote. The only thing that got me really thinking about this is that when it comes to "bonding" with the charge field -- essentially creating complex molecules, Graph databases may allow ...
Airman. Cr6, granted. I agree, a graphDB would likely provide alternative coding possibilities that should, for example, make bonding solutions more direct and natural than when using Relational data tables. I’ve been stuck in a rut, anxious, and hoped for some needed discussion which you've kindly provided. Thanks.
A blast from the past.
Airman. A quick search shows Jupyter-Databricks is defined as an Apache Spark analytics platform, separate from the current graphDB reviews and discussion, but still something to keep in mind.
Post on this thread by Chromium6 Sun Jan 23, 2022 11:00 pm
Hi Guys,
I uploaded a slightly modified MathisPeriodicTable .csv with column header names changed so that works with Jupyter-Databricks:
mediafire.com … MathisPeriodicTable.csv
import pyspark
from pyspark.sql import SparkSession
from pyspark.sql.types import *
import pyspark.sql.functions as F ...
The best argument for using a graphDB is your larger MathisPeriodicTable.csv excel spreadsheet with 157 columns. You assembled a great deal of pertinent information that has not been included yet. At the time I was trying to learn pandas and said we would need separate interfaces for each of the data columns – many custom widgets. Oh, and Lithium is missing.
As I now see it, a graphDB (or ipycytoscape or networkX) graph network can allow each atomic vertex/node, or edge to contain most all those additional property data values, resulting in extremely feature-rich graphs. Maybe by adding a single additional graph interface, a widget or IPython display may allow interactive displays of all that additional data. Such an interface could be used to launch model selections as you have described. I suppose the current interactive Periodic table widget could be replaced with an expanded interactive graph diagram, one that might include a detailed display of the atom currently highlighted. Such an interface could allow access to most any of the 157 column data.
And another thing, as per our previous discussions, understandings and/or agreements, before we go throwing arbitrary molecules about, we’ll need to try incorporating machine learning. You’ll no doubt tell me ML is quicker and easier with feature rich data rather than many relational table data look-ups. By then we should have a graph with ML capabilities.
Cr6 you mentioned extensive coding. By that I take it to mean converting all the relational table data into graph data. Much as I’d like to, I’m clearly not making any progress on molecules front at present. I’d be more productive learning how to convert the .csv or pandas table data into graph form. I’ll see if I can create any graph versions of AtomBuilder3 starting with NetworkX.
Sound Ok to you?
Pretty please, could you make your larger, unmodified MathisPeriodicTable.csv file available again?
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
LTAM wrote:I believe the following are all 8 possible e-p TB RL slot configuration types the program can now create. Each type may come in any one of five, 2–6, e-p versions.
8 possible (e-p)/(e-p) bond types and each type’s 2-6 (e-p) proton configurations.
https://tinkerpop.incubator.apache.org/docs/3.0.1-incubating/#traversal
.csv importers:
https://github.com/vsantosu/gremlin-importer/wiki/CSV-import-guide
https://openbase.com/js/gremlin-importer/documentation
https://db-engines.com/en/system/TinkerGraph%3BTitan
Besides TitanDB-JanusGraph there is SageMath that has math libraries for Python-Jupyter notebooks.
https://github.com/thinkaurelius/titan/ (retired)
https://janusgraph.org/ (current updated titan version)
SageMath (has all libraries we might need and runs as a jupyter server)
https://github.com/sagemath/sage-windows/releases
Last edited by Chromium6 on Mon Oct 03, 2022 1:18 am; edited 3 times in total
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
https://www.mediafire.com/folder/7uqcs0d5l0cs0/Mathis
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

A SlotLayout level graph representation showing 19 slot nodes and 18 edges - the bonds between slots.
- Code:
import networkx as nx
import matplotlib.pyplot as plt
G = nx.Graph()
G.add_nodes_from([
(1, {"slot_axis": "Z"}),
(2, {"slot_axis": "Y"}),
(3, {"slot_axis": "Y"}),
(4, {"slot_axis": "Z"}),
(5, {"slot_axis": "Z"}),
(6, {"slot_axis": "Y"}),
(7, {"slot_axis": "Y"}),
(8, {"slot_axis": "X"}),
(9, {"slot_axis": "X"}),
(10, {"slot_axis": "Z"}),
(11, {"slot_axis": "Z"}),
(12, {"slot_axis": "Z"}),
(13, {"slot_axis": "Z"}),
(14, {"slot_axis": "Y"}),
(15, {"slot_axis": "Y"}),
(16, {"slot_axis": "Z"}),
(17, {"slot_axis": "Z"}),
(18, {"slot_axis": "Z"}),
(19, {"slot_axis": "Z"}),
])
slot_edge_list = [(1, 2), (1, 3), (1, 6), (1, 7), (1, 8), (1, 9),
(2, 4), (2, 16), (2, 18), (3, 5), (3, 17), (3, 19),
(1, 9), (4, 14), (5, 15),
(6, 10), (7,12), (8, 11), (9, 13)]
G.add_edges_from(slot_edge_list)
nx.draw(G,with_labels=True)
plt.savefig("SLGraph2.png")
No problem, Cr6, except that I jumped right into networkx after my last post. Networkx.org provides a starter tutorial. I finished that and began building another tutorial from the documentation pdf.Cr6 wrote. Sorry for delaying an update. I did more research on this. … .
https://networkx.org/documentation/stable/tutorial.html
https://networkx.org/documentation/latest/_downloads/networkx_reference.pdf
https://networkx.org/documentation/stable/auto_examples/index.html
Here’s a quote from near the top of the tutorial.
I’ve also watched additional networkx videos.By definition, a Graph is a collection of nodes (vertices) along with identified pairs of nodes (called edges, links, etc). In NetworkX, nodes can be any hashable object e.g., a text string, an image, an XML object, another Graph, a customized node object, etc.
So far, networkx is much easier to learn than pandas. The above graph diagram and code has 19 slot nodes, and 18 slot bonds indicated by edges. Of course the diagram is not intended to display an image of a real atom. Only a representation, of the relationships, properties and/or attributes – between slots and slot bonds.
Not sure what you mean, it seems people design graphs in one of two forms, node-centric or edge-centric. I’m node centric, nodes are objects. Above is is a graph of the the slotlayout level. Each slotlayout level node is its own graph (or will be anyway) representing the 0-6 proton groups (a proton group is the lowest level e-p-n sub graph, also not yet included) contained in that slot. The lines kind of suggest and may be interpreted as charge channel bonds between slot centers.Cr6 wrote. We may be able to create edges for Slots and e-p protons with the larger file?
As with the rest of the program, I’m essentially basing things on the slot layout diagram, not to imitate reality, but as a good starting point. The many many data attributes or properties in your larger csv file will add rich feature dimensionality to whatever level node or edge those properties are attached to.
You’d likely build an atomic graph differently, please share it if you do. As of yet, the only attribute included in the code so far is in the slotlayout graph shown, each node has a "slot_axis", property with X, Y or Z entries. The TBRL e-p-n position/spin rules may need to be node attributes of both the slotlayout and the slot level graphs. Edge attributes may include spins, positions and distances.
NetworkX appears rich in pre-built algorithmic graphs, but it may turn out to be deficient in its analytic capabilities – i.e. in identifying atomic bonding sites. Unless you have any objection or direction otherwise, I think I’ll continue playing with networkx for the time being.Cr6 wrote. I'm leaning towards TinkerPop3 only because there are more examples available for programming with it. Neo4J is easier to use but limited unless you buy it.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Sorry Cr6, not much to report. I was able to find and follow along a few more pandas dataFrame/networkX examples.
https://networkx.org/documentation/stable/reference/convert.html
According to the book, (read "pandas" as "dataFrame") the preferred way of converting data to a NetworkX graph is through the graph constructor. The constructor calls the to_networkx_graph() function which attempts to guess the input type and convert it automatically.
to_networkx_graph. ...Container (e.g. set, list, tuple) of edges iterator (e.g. itertools.chain) that produces edges generator of edges Pandas DataFrame (row per edge) 2D numpy array scipy sparse matrix pygraphviz agraph create_usingNetworkX graph constructor,...
For convenience, There seems to be two functions that are used to convert from a pandas dataFrame to a networkX graph.
1. from_pandas_adjacency
...from_pandas_adjacency from_pandas_adjacency(df, create_using=None)[source] Returns a graph from Pandas DataFrame. The Pandas DataFrame is interpreted as an adjacency matrix for the graph. Parameters: dfPandas DataFrameAn...
2. from_pandas_edgelist
...source='source', target='target', edge_attr=None, create_using=None, edge_key=None)[source] Returns a graph from Pandas DataFrame containing an edge list. The Pandas DataFrame should contain at least two columns of node names and zero or mo...
And two functions to convert from a networkX graph to a pandas dataFrame
1. to_pandas_adjacency
2. to_pandas_edgelist
As of yet, it still makes little sense. Although it now seems likely I’ll need to somehow restructure the atomic slot layout pandas datafame in order to conform with networkx’s node-to-node edge format.
Also I need to forget nesting nodes for the time being, if not longer. No collapsing or expanding graphs. All the electrons, neutrons and protons for any given atom will be part of a single atom graph.
All’s well. I’ll keep at it.
.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
https://www.datanami.com/2022/10/05/overcoming-data-integration-challenges-to-unleash-massive-potential-knowledge-graphs-101/
Chromium6- Posts : 735
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Thanks Cr6. I agree, that's a good recap. The 30,000 ft assessment including data silos. Some practical considerations intended to improve an enterprise mission’s success – how best to deal with data changes before they happen. I can mostly understand and appreciate the advice, and almost hear your voice Cr6. Thanks again for your guidance. If you gave more explicit detailed instructions I might be able to follow better. As it is, all things including ML taken into consideration - granted, a knowledge graph may well be the only way to go.
Since a knowledge graph is a more sophisticated form of network graph, I think I still need to learn simple network graphs first. That is, NetworkX. Between you and me I broke down and purchased an introductory study guide – Working with Networks in NetworkX. Registering the purchase should have entitled me to a free copy of all the files used in the book, but the registration direction is out of date and I stopped short of purchasing an electronic copy. They do a good job at explaining the easy stuff, better than the documentation. So far so good concentrating on networkX's built-in karate club function data. I expect missing files will likely prevent me from working through the whole book, less than 180 pages. Hopefully I’ll understand the networkX documentation better by then too. Shouldn’t take too long. We may or may not want to end up with a networkX version of the project, but if we did I’d consider that good progress. Maybe then attempting the more complicated knowledge graph will make more sense.
That’s just the sort of thing your source advises against, locking in some sort of data structure that eventually needs changing which greatly complicates things in the long run. Choose you data wisely. At best, redirecting enterprises delays any possible success, etc. but I don’t think the sort of zig zagging efforts your source advises against is necessarily a problem for us. Unfortunately or not, this is a learning process. Given my level of knowledge and ability I’d say the time and effort spent in remodeling a project again and again is a good thing - refinement. Take your pick. For example, Pandas was a great addition, allowing even more possible changes to the project and in no way a waste of time, nor does it necessarily lead to a knowledge graph.
Please let me know if my overall lack of progress and general lackadaisical attitude has caused you any grief.

.
LongtimeAirman- Admin
- Posts : 2034
Join date : 2014-08-10
Chromium6 likes this post
Page 4 of 12 •  1, 2, 3, 4, 5 ... 10, 11, 12
1, 2, 3, 4, 5 ... 10, 11, 12 
 Similar topics
Similar topics» Cr6's Periodic Table
» RSC's Periodic Table for Comparison
» Happy sesquicentennial to the periodic table of the elements
» Periodic Table of Toxic Elements