Miles Periodic Table with Standard Periodic Table reference
Page 10 of 12 •  1, 2, 3 ... 9, 10, 11, 12
1, 2, 3 ... 9, 10, 11, 12 
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Cr6 wrote. Just wanted to get your eyes on this query. It is kind of "check" on what is allowable. Looking for unique or low counts for src-dest with the LTAMKeys and CanBind = 1 (true). There are 2,500 + unique values. Was looking at A1 through A3 LTAMKeys. Trying to firm up "allowed" and "not-allowed:"
- Code:
With SmallBondCounts as (
SELECT TOP 100 PERCENT
LTAMKeySrc
,LTAMKeyDest
...
--------------------
This shows a 1 Record count. This Formula has Barium and Radium bonding with unique LTAM Keys:
- Code:
SELECT *
FROM
[Physics].[dbo].[vwAtomicMilesMathisOrbitalsDetailAllBonds]
where
LTAMKeySrc = 'A3Y6A' and LTAMKeyDest = 'B10Z5R'
Airman. Copy that Cr6. If I understand correctly, all molecular bonds can be identified, enumerated, tabulated and counted. The SmallBondCounts query identifies the molecular bonds that occur the least amount of times. Likely identifying problems, definitely useful in checking and/or validating the bonding logic. I’m surprised there are at least 2,500 unique bonds. I would think they should all be verified. Its likely most of the problems found would apply to more than a single bond.
ZID Formula
29315100 BaO8RaS2
-------------------
Just wanted to get this checked out for validity.
Ok, looking at the single occurrence: LTAMKeySrc = 'A3Y6A' and LTAMKeyDest = 'B10Z5R' which can occur for the bond between barium and radium within the molecule BaO8RaS2.

The positioning of the two scatter plots and the double sided red arrow between them is intended to align and show Ba hook slot3, Y-Z m-f bonded to the center (negative y side) and outer edge of Ra’s carousel slot10. This exercise makes it clear it may be time to add additional atoms plots to the notebook - and I need to get back to graphs.
Yes, Ba can bond at either end of slot3, but I believe those two possible bonding sites - Ba’s negative z hook positions, would be ‘filled’ if they are occupied with small atom, single epn stacks consisting of 1, 2 or 3 protons, and not the main side-to-side charge channel of a spinning carousel, containing 12 direct in-line protons sharing the same spin axis, charge channel, separated by three orthogonal z axis slots.
Yes, I allow that Ra can bond at the end of its main side-to-side carousel spinning charge channel, but if it did, I imagine the carousel would no longer be spinning. I believe a bond at at the outside edges of the carousel could indicate a non-spinning carousel, as might be present in some sort of crystal molecule.
Until we have rules covering crystals I suppose the ‘rules’ need to be clarified to disallow that particular Ba-Ra bond.
Your SmallBondCounts query result showed that there are 5 additional combinations of Ba and Ra source and destination keys which also occur only once among all molecular bonds:
No. Formula ElementSrc ElementDest LTAMKeySrc LTAMKeyDest RecCount
1 BaO8RaS2 barium radium A3Y6A B10Z5R 1
2 BaO8RaS2 barium radium T9X6L B10Z5R 1
3 BaO8RaS2 barium radium A3Y6A T12Z5L 1
4 BaO8RaS2 barium radium V2Y6V B10Z5R 1
5 BaO8RaS2 barium radium T6Y6L B10Z5R 1
6 BaO8RaS2 barium radium V2Y6V T12Z5L 1
No. 1 is described as disallowed above.
No. 2, ‘T9X6L’ and ‘B10Z5R’ would indicate that the outer carousel edges of both atoms may be joined, as, I believe, may be allowed for a crystal.
No. 3, ‘A3Y6A’ and ‘T12Z5L’ is the same alignment as no.1 but with Ra rotated 180deg about its z axis, and could be disallowed for the same reasons as no.1.
No. 4, ‘V2Y6V’ and ‘B10Z5R’ is between one of Ba’s positive z hook positions and the outer edge of Ra’s other main side-to-side charge channeling and usually spinning carousel. That bond should also be disallowed for the same reasons as no.1.
No. 5, ‘T6Y6L’ and ‘B10Z5R’ is much the same as no.2 but with Ba rotated 90deg – a possible crystal bond.
No. 6, ‘V2Y6V’ and ‘T12Z5L’ is between Ba’s positive hook position and a 180deg rotated Ra, and should be disallowed for the same reason as no.1.
Two possible valid crystal bonds and four disallowed hook position bonds. Do you agree?
Again, I'd argue for slot key variability. With alternate possible rotations, bonds can be very flexible, but if an explicit key assignment is incorrect, we might be missing important assignments and their resulting bonds.
///\\\\/////\\\\\\/////\\\\///\\
Airman. Given ep and ps column data values: ‘T’ (1), ‘B’(-1), ‘L’(-1), ‘R’(1). I guess a third value zero null placeholder is exactly what a zero in -1,0,1 would be - the lack of either +/-1 values.From SQL and the Snare of Three-Valued Logic by Joe Celko.
The whole subject of the Three-Valued (also known as ternary, trivalent or 3VL) Logic of SQL tends to trip people up. This is hardly surprising in view of the fact that it involves an esoteric Polish mathematician and because it behaves differently in the DDL (Data Declaration Language) and the DML (Data Manipulation Language). In response to requests, Joe Celko comes to the rescue and makes it all seem simple.
...
Here it is! But the problem is that SQL has a three valued logic (3VL) and not the two values of Mr. George Boole. In fact, SQL logic is complicated.
...
Like many problems, this starts with NULLs. Dr. Codd put the concept into the first version of the Relational Model. It is not a value; it is a “place holder” for a value.
The snare in the title is cautionary but if SQL uses 3VL such as when including a zero null value, and not ‘binary boolean logic’, then I guess I should try to learn the difference.
After a single read the SQL 3VL subject matter seems hopelessly esoteric and complicated. It may have no bearing, but must admit I liked my Hewlett-Packard scientific calculator and preferred its reverse Polish key entry sequences over the Texas Instruments usual or ‘standard’ calculator key entries. HP calcs were cleaner and faster, with no need to press an equal key.
To Be Continued ...
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
https://www.chemspider.com/Chemical-Structure.29315100.html?rid=9efd0b90-f966-4e06-9544-a49728d4bba4
Barium radium sulfate (1:1:2)
Molecular Formula BaO8RaS2
From what is loaded thus far... it is stand alone apparently:
SELECT *
FROM [Physics].[dbo].[vwAtomicPivoted]
Where (AtomicSymbol = 'Ba' or AtomicSymbol = 'Ra')
and (ZFormula like '%Ba%Ra%' or ZFormula like '%Ra%Ba%' )
Order by ZFormula, MolOrder2
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
SQL 3VL. Upon further reading, Joe Celko’s SQL and the Snare of Three-Valued Logic continued to frustrate me. Fortunately, a search on SQL 3VL returned many additional descriptions and discussions. At Modern SQL, The Three-Valued Logic of SQL by Markus Winand https://modern-sql.com/concept/three-valued-logic contained many clear logic explanations and examples which made good sense and restored my well-being. I may not be any closer to determining whether matricies can be used to determine bonds in the ‘Physics’ database, but I do understand that the presence of null values within SQL databases adds complexity and 'unknown' boolean results when comparing database values.
Airman. I read and obey Cr6.Cr6 wrote. https://www.chemspider.com/Chemical-Structure.29315100.html?rid=9efd0b90-f966-4e06-9544-a49728d4bba4
Barium radium sulfate (1:1:2)
Molecular Formula BaO8RaS2
I just noticed your post also includes:
Airman. I've not taken that stand alone fact or select query into consideration.From what is loaded thus far... it is stand alone apparently:
SELECT *
FROM [Physics].[dbo].[vwAtomicPivoted]
Where (AtomicSymbol = 'Ba' or AtomicSymbol = 'Ra')
and (ZFormula like '%Ba%Ra%' or ZFormula like '%Ra%Ba%' )
Order by ZFormula, MolOrder2
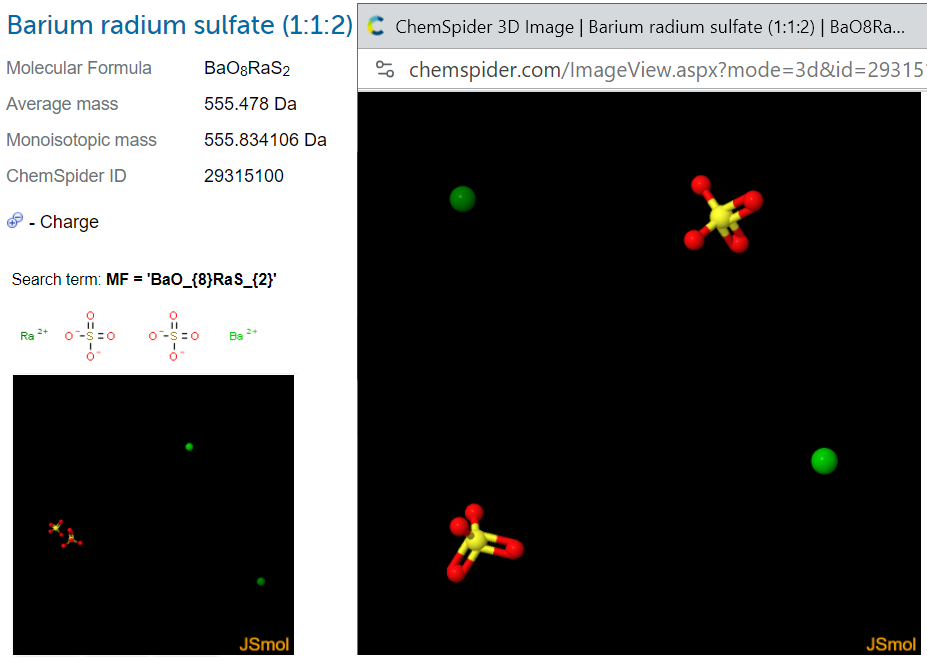
Barium Radium Sulfate. Here are two different 3D views (and sizes) of Barium Radium Sulfate, BaO8RaS2 from chemspider.com. Nice to see the 3D view can be zoomed and rotated about. Apparently Barium and radium are the blue and green spheres - without any bonds? The only bonds I see are between each sulfur and four surrounding oxygen atoms. Doesn’t look like a crystal or even a molecule to me; or anything like what I posted about last time. The 2D graph(?) also seems insufficient, only showing bonds for the molecule’s two SO4 groups(??).
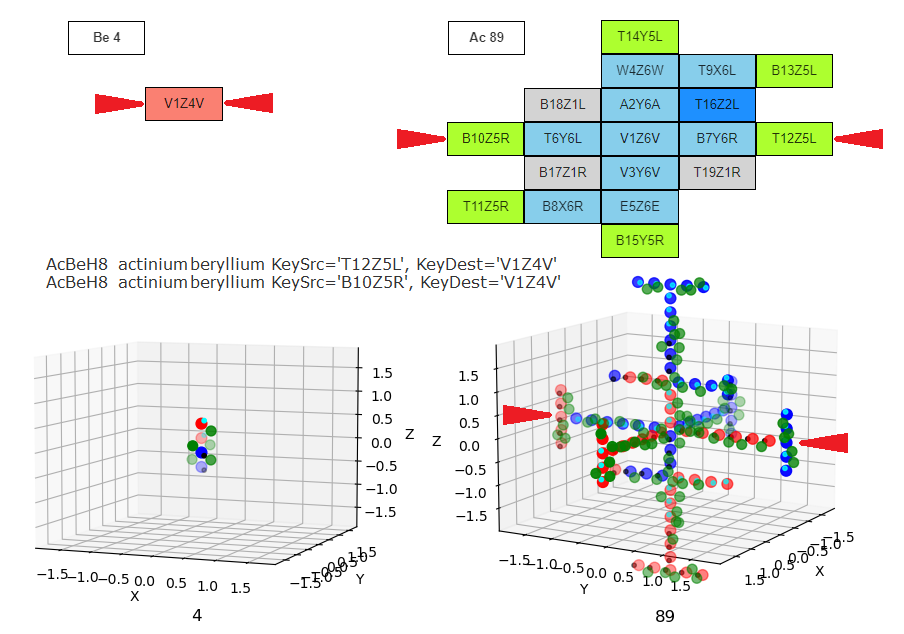
SmallBondCounts query outputs for AcBeH8 and ThBeH8. Returned to the SmallBondCounts query output in order to try some more charge bond verifying. This time for four RecCounts, 2 bonds between actinium and beryllium and 2 identical bonds between thorium and beryllium:
Formula ElementSrc ElementDest LTAMKeySrc LTAMKeyDest RecCount
AcBeH8 actinium beryllium T12Z5L V1Z4V 2
AcBeH8 actinium beryllium B10Z5R V1Z4V 2
Formula ElementSrc ElementDest LTAMKeySrc LTAMKeyDest RecCount
ThBeH8 thorium beryllium T12Z5L V1Z4V 2
ThBeH8 thorium beryllium B10Z5R V1Z4V 2
SmallBondCounts output column headers:
ZID Formula CurrentAtom NextAtom1 NextAtom2 CurrentAtomCount NextAtom1Count NextAtom2Count LTAMKeySrc LTAMKeyDest AtomicNumberSrc AtomicNumberDest SlotDirectionElectronBond IsValid
Two SmallBondCount outputs for LTAMKeySrc = 'T12Z5L' and LTAMKeyDest = 'V1Z4V'.
12345 AcBeH8 Ac Be H 1 1 8 T12Z5L V1Z4V 89 4 TLVV 1
12346 ThBeH8 Th Be H 1 1 8 T12Z5L V1Z4V 90 4 TLVV 1
Two SmallBondCount outputs for LTAMKeySrc = 'B10Z5R' and LTAMKeyDest = 'V1Z4V'.
12345 AcBeH8 Ac Be H 1 1 8 B10Z5R V1Z4V 89 4 BRVV 1
12346 ThBeH8 Th Be H 1 1 8 B10Z5R V1Z4V 90 4 BRVV 1
First, I should mention that the beryllium atom shown in the plot above and described by the slot1 TBLR code of V1Z4V is not the form of Be described by Miles and included in our original Cr6-Elements.csv atomic data – for which Be’s four protons occupy three slots: 2 protons in slot1 and 1 proton each in both slots 2 and 3. That discrepancy doesn’t seem to be an issue for discussing the SmallBondCounts outputs in AcBeH8 and ThBeH8.
Be is a four proton stack and both Ac (and Th) slots10 and 12 are five proton stacks. Nine protons is greater than the maximum number of protons allowed for a shared slot bond, so the resulting bonds must be Y-Z m-f types. Be is initially oriented along the z axis, but it can rotate 90 degrees about the X axis – changing to a Y orientation, and so could be expected to form a Y-Z m-f bonds with Ac (or Th) slots 10-13.
When I look at the SlotDirectionElectronBond data I see TLVV for Be bonded at Ac (or Th) slot 12, and BRVV for Be bonded at Ac (or Th) slot 10. Since ‘V’ is a TRBL configuration TLVV and BRVV are not valid. If Be slot1 was a ‘A’ type, A1Z4A then TLAA and BRAA would be valid. Since ‘V’ is a TRBL configuration, I would expect Be and Ac (or Th) slots 11 and 13 to form valid bonds with a TRVV and BLVV. That logic would be appropriate for a resulting shared slot bond. We don’t have any such detailed TBLR bonding rules that apply to Y-Z m-f bonding. Be atoms bound to the outer edge of an Ac (or Th) carousel would likely unbalance the carousel. I'm probably missing a few details, feel free to point them out. Any comments?
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Cr6, sorry, as usual, thinking about it, I see places where I’ve been wrong. We may have enough ortho slot bonding rules.
First, my m’s and f’s with respect to stack ends (what I previously referred to as male) or stack centers (what I've called female) describing ortho bonding are opposite Miles’ definitions of m’s and f’s, where a proton’s equatorial emissions (male - center stack) feed directly into an adjacent, orthogonally oriented proton pole (female - stack end). Below and in the future I will refer to male and female ortho stacks as per Miles' definition.
Secondly, I believe the T/B electron rule that explains bonding between vertical spin axis aligned protons, which can only occur due to the low charge pressure zone caused by an electron occupying the proton’s opposite pole, can also help account for bonding between adjacent orthogonal epn proton stacks. One or both orthogonal epn stacks must contain one or more low charge pressure zone.
Single spin, single electron position stacks (TRTR, TLTL, BRBR, BLBL ) have a single low charge pressure end without an electron. By definition, TRTR and TLTL type stacks form low charge pressure zones at those stacks’ bottom ends, and BRBR and BLBL type stacks have low charge pressure top ends. Those ‘female’ proton stack ends may bond as per Miles definition, with another stack’s center proton emissions feeding the orthogonally adjacent and I would add - electron free, low charge pressure proton pole.
Then I hit a problem. Both ends of T/B alpha stacks TLBR, TRBL, TRBR, and TLBL contain electrons at both top and bottom stack ends and so cannot form low charge pressure zones an their stack ends. It occurred to me that there would be no problem if T/B alpha stacks can ortho bond due to the low pressure zone present inside the stack’s central alpha. In which case I believe that when two alpha stacks get together, they can ortho bond at either stacks’ low charge pressure center and not to an off-center stack’s single proton emission field. I believe that bond that can form despite adding the ortho stack's T or B electron to one side of the 'male' alpha’s stack's center.
R or L proton spins? Other than to allow only the eight TBLR type stacks, or to somehow maintain a general right hand rule between ortho bonded stacks, I don’t see how R or L proton spin changes would effect the main T/B electron position and Miles’ m/f rationale above.
If they’re Ok with you, I believe these ortho stack bonding rules should be sufficient for all our needs. I don't see any existing TBLR assignment problems yet, but I may or may not need to make some assignment changes in accordance with them.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
LTAM wrote:Cr6, sorry, as usual, thinking about it, I see places where I’ve been wrong. We may have enough ortho slot bonding rules.
First, my m’s and f’s with respect to stack ends (what I previously referred to as male) or stack centers (what I've called female) describing ortho bonding are opposite Miles’ definitions of m’s and f’s, where a proton’s equatorial emissions (male - center stack) feed directly into an adjacent, orthogonally oriented proton pole (female - stack end). Below and in the future I will refer to male and female ortho stacks as per Miles' definition.
I see a bit at what you are getting at. No worries...we can have different databases with different configurations if necessary like DEV, QA, PROD style. With this type of matchup between Miles and somewhat documented known formulas there needs to be a window of "discovery-trial/error" and this may be on-going for awhile. The
"strange ones/outliers/singletons" might need the most attention for awhile.
BTW, this is a good paper to re-read: http://milesmathis.com/magnus.pdf
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Cr6 wrote. I see a bit at what you are getting at. No worries...we can have different databases with different configurations if necessary like DEV, QA, PROD style.
Airman. Thanks, Cr6, “No worries …” is a relief. The idea of different databases had never occurred to me. For better understanding I google searched on “DEV, QA, PROD”. The top result:
Development and Production Database?
https://stackoverflow.com/questions/299986/development-and-production-database
Without getting into all the details, on Nov 18, 2008 at 20:16, Humpton mentioned his appreciation of source control and basically asked how he should proceed in further developing his database project. Just three minutes later, ahockley answered with the top ranked reply:
That and other similar returns make good sense. Taking note that “all database objects should be scripted and stored in source control” - the next thing to wonder about. https://stackoverflow.com/questions/5731969/sql-server-source-controlOn Nov 18, 2008 at 20:19 ahockley wrote.
Each environment should have a separate database. Script all of the database objects (tables, views, procedures, etc) and store the scripts in source control. The scripts are applied first to the development database, then promoted to test (QA, UAT, etc), then production. By applying the same scripts to each database, they should all be the same in the end.
If you have data that needs to be loaded (code tables, lookup values, etc), script that data load as part of the database creation process.
By scripting everything and keeping it in source control, a database structure can be recreated at any time for any given build level.
Unless you’ve made a great deal of progress I’m unaware of, I suppose ‘Physics’ is currently a single developmental environment. Then all the tables, views, stored procedures and such constitute an initial source control. Thanks for all your unremarked and unrecognized hard work and guidance.
Cr6 wrote. With this type of matchup between Miles and somewhat documented known formulas there needs to be a window of "discovery-trial/error" and this may be on-going for awhile. The "strange ones/outliers/singletons" might need the most attention for awhile.
Airman. Copy that Cr6, an SQL database can be complex stuff, glad you know what you’re doing. “Strange ones/outliers/singletons” sounds like the place for me.

Strange ones/outliers/singletons. Here are the next three single occurrence, SmallBondCount bond outputs – this time for AgF4Xe2. In all three cases Xenon’s bonding site is identified as its z oriented top, slot4, ‘W4Z6W’ or ‘TLTLTLBLBLBL’. The top of slot 4 consists of an electron occupied proton pole which prevents the stack from being a low charge field pressure zone. The three identified F-Xe bonds occur at each of F’s three available slots: a. F slot3, ‘B3Y2R’ or ‘BRBR’; b. F slot1, ‘V1Z6V’ or ‘TRTRTRBLBLBL’; and c. F slot2, ‘T2Y1L’ or ‘TR’.
a. Xe ‘W4Z6W’ and F ‘B3Y2R’ is not a valid molecular bond. By the rules I identified in my last post, the only type of slot that can bond with Xe’s +z electron end is a T/B alpha type stack containing a low charge pressure stack center, such as: ‘TLBR’, ‘TRBL’, ‘TLBL’ or ‘’TRBR. F slot3 has another problem – an invalid atomic bond between F slot1 and F slot3. This time, Fluorine’s atomic slot3 data must be changed from the current ‘BRBR’ type to any of the T/B alpha types to correct the atom.
b. Xe ‘W4Z6W’ and F ‘V1Z6V’ looks like a valid bond to me. A criticism might be – why are the neutrons in the way? Yes, I agree, I think they need to be closer together.
c. Xe ‘W4Z6W’ and F slot2, ‘T2Y1L’. I do not believe a single epn proton set can provide a valid molecular bond between two full orthogonal T/B alpha type stacks. Like F's problem slot3, the current F slot1 to F slot 2 bond is an invalid atomic bond. It looks like F slot1 needs changing first.
That’s all for now. I'll review http://milesmathis.com/magnus.pdf next.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Individual spinning protons constantly recycle charge photons which enter the proton mainly through both of the proton’s spin axis poles. Bonds between protons can only occur when each spinning proton has a captured electron circling either the proton’s top or bottom pole. The electron blocks charge from entering that proton pole which creates a low charge pressure zone near the proton’s other pole. In Diatomic Hydrogen *, Miles explains all that, and identifies eight valid bond combinations between vertically adjacent vertical spin axis aligned protons (spinning right or left), and their attendant electrons (at either the proton’s top or bottom pole).
All the protons within each valid stack of ep sets share a single common charge channel. As each ep is added, the amount of charge passing through all the protons channel is increased. The maximum number ep sets is six. A seventh proton can still be bound to the stack’s relatively low charge pressure (electron-free) end, but the combined six proton charge channel flow overwhelms and turns the seventh proton’s orientation 90 degrees, which blocks and redirects some the charge throughput it receives and recycles. The seventh proton cannot be a member of the six proton stack; the seventh proton must be part of a second 1-6 ep set stack. Thus two different types of ep bonding have been described: 1. parallel/anti-parallel bonding that occurs in a single shared stack; and 2. orthogonal bonding that occurs between adjacent, orthogonal ep stacks, each consisting of 1-6 ep’s.
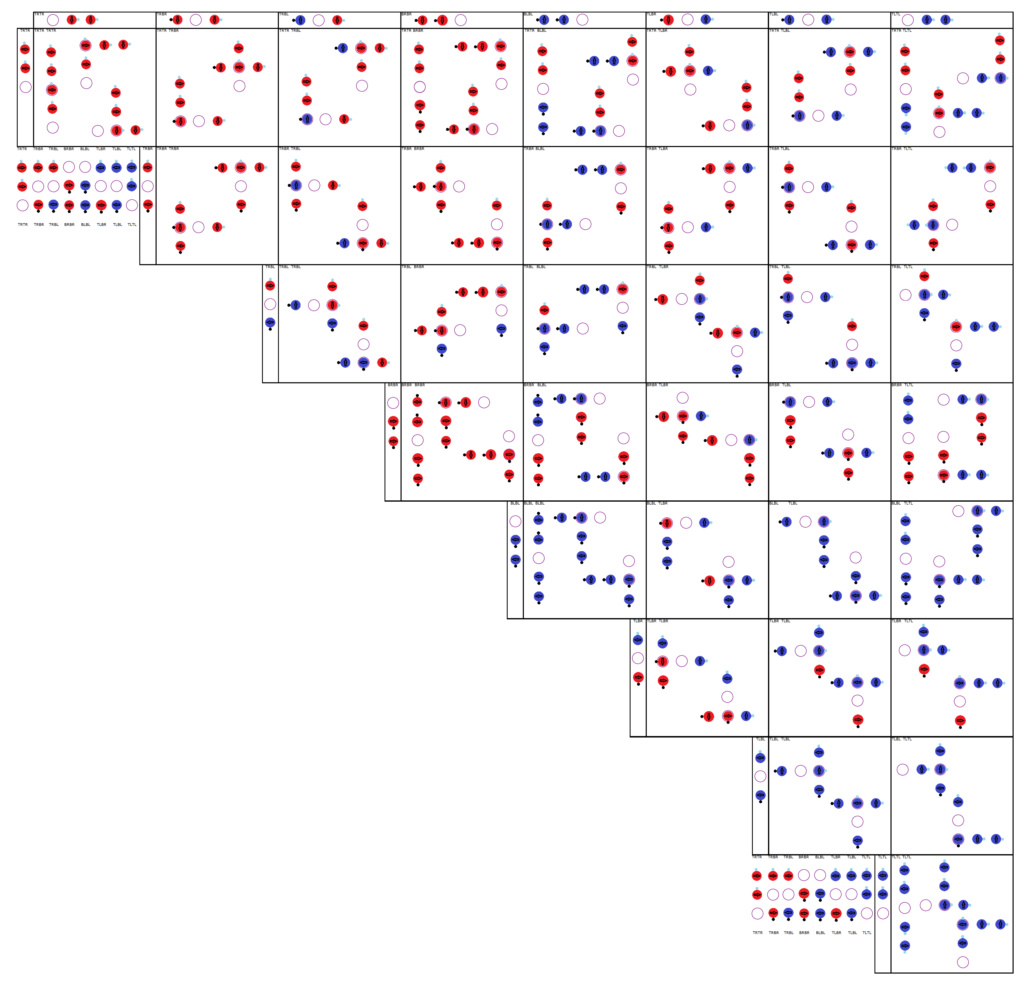
The above diagram is large and busy, and please pardon my usual errors. For starters it includes not one but two diagrams of the eight allowed, valid ep stack types for easy reference. I’ve added low charge pressure zones – violet circles, where I believe ortho bonding can occur, one for each of the eight valid ep stack types. Thirty-six panels contain mostly two examples of four total possible, adjacent stack, orthogonal bonds; each of the eight types may bond with each of the eight types that have been rotated 90 degrees clockwise. Each panel’s two ortho examples shows one of the protons of the second bound within the low charge pressure zone of the first set, and a vise versa example where first set occupies a low pressure zone of the second set. I believe each panel contains two of the four possible bond alternatives. Each stack contains only two ep sets, but I also believe the same rules apply to two adjacent stacks of 1-6 ep’s. Ten panels include a separate set of in-line bonds which are not a part of the ortho bonding 90 CW degree rotation set.
The main difficulty I’ve been having is in attempting to visualize the alpha type stack’s central low pressure zone. The ten panels including a not 90 degree rotation contain examples that do not involve alpha type central low pressure zones. I may be wrong but I don’t believe that just those 10 panel type ortho bonds could describe all ortho bonds. Another thing, the alpha center has no central vertical charge channel in-line bottom or top electrons, yet it appears single electrons are allowed in the alpha zone’s other two, intersecting ortho charge channel proton pole positions. I keep wondering how the orthogonal charge channels within that low pressure volume of space might pass, intersect or interact. And what about the neutrons? How does everything fit? In other words, I still haven’t reached a real definitive/possible alpha zone understanding yet.
*
http://milesmathis.com/index.html
332. Diatomic Hydrogen. My new charge bonding explains this much better than electron sharing. Plus an analysis of spin isomers. 9pp. http://milesmathis.com/diatom.pdf
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

The diagram includes thirty-six panels, each including four possible combinations of orthogonal bonds between adjacent vertical and 90 degree cw rotated, electron/proton (ep) stacks. Each stack contains two aligned ep sets as well as a single low charge pressure area - indicated by a violet circle, where a proton of a second stack may orthogonally bond to the first. A total of 144 possible orthogonal bonds.
Airman. Glad you think so Cr6. With that kind of high praise I decided I better continue working it, to make it as clean as I could. Changes include enlarging it to fit each panel’s four combinations, dropping the 10 non-orthogonal in-line entries and adding the remaining 72 possible diagrams. Rotated one of the two ‘eight valid stack’ reference diagrams 90 degrees cw and corrected a few errors. Hope I haven’t blunted any ‘cool edginess’. Please let me know if you think it needs any further corrections or changes, or any other requests. I’m especially intrigued by your mention of related notebook changes(?!?).Cr6 wrote. Nice graphic LTAM! Helped me see the alignments in a new perspective. I could work on adding a few via jupyter notebooks. May need a formula drop down widget that could try to show layout. Your graphic looks "edgy" and frankly "cool". I'm still dorking around with orientdb for graph style bidirectional bonding...but I'm seeing similar issues with showing atom to atom bond relationships. I can show that this formula "forms" with this binding according to Mathis....but to really show "why" is still tricky. You are taking that step though!
You requested - if at all possible, matrices; I’m not sure if this graphic complies. Do you agree that T/B alphas can support internal orthogonally oriented protons and proton stacks extending out from the alpha’s central void?
I wonder how the data shown might be reduced to some sort of formula?
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
LTAM wrote:
Please let me know if you think it needs any further corrections or changes, or any other requests. I’m especially intrigued by your mention of related notebook changes(?!?).
You requested - if at all possible, matrices; I’m not sure if this graphic complies. Do you agree that T/B alphas can support internal orthogonally oriented protons and proton stacks extending out from the alpha’s central void?
I wonder how the data shown might be reduced to some sort of formula?
I don't see why a T/B configuration like that would not be valid. Maybe Miles has a quote or example somewhere to disallow it? Sounds reasonable as you have described it.
Reducing this to a formula could get interesting. Especially if it could allow for atom bonding from it. Would be an interesting challenge.
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

A possible update to the valid ep stack diagram. Identifying the fact that T/B alpha stacks (TRBR, TRBL, TLBR, TLBL) contain two low charge pressure zones, due to both those stacks’ top and bottom ep’s.
Cr6 wrote. “I don't see why a T/B configuration like that would not be valid. Maybe Miles has a quote or example somewhere to disallow it? Sounds reasonable as you have described it.”
Airman. Thanks for the reassurance Cr6, I’ll try describing a problem or two.
Throughout this project and earlier I’ve assumed any given atom’s spinning carousel center – slot1, could be a z-aligned alpha; my understanding of alphas has developed in the meantime, mostly according to my more recent re-interpretation of Diatomic Hydrogen *. I don’t recall any Miles quote or example disallowing or even mentioning any alpha center orthogonal bonding. I believe I would remember if he had but I’d be happy to be proven wrong. To the best of my knowledge, Miles has never placed orthogonal protons inside alphas, only neutrons.
Its entirely reasonable that bonding for both in-line and adjacent orthogonal stacks can only occur at low charge field pressure zones (low-cfz’s), near the proton pole opposite the electron occupied proton pole. Your term - “a T/B configuration like that”, is a positive, reasonable and agreeable hedge since we don’t know the details.
As the above ‘updated’ valid ep stack diagram is intended to show, a T/B alpha type ep stack configuration has two possible vertically aligned low-cfz’s; one is created below the alpha’s top ep set and the second is created above the alpha’s bottom ep set. In my previous 144 ortho bonds graphic I replaced the two with a single central low-cfz O without special mention or further discussion. That’s likely a mistake, I’ll elaborate.
There are two separate vertically aligned potential low-cfz’s within a T/B alpha. What would keep the two low-cfz bound protons from drifting together? Sure, protons emit charge photons in all directions but mainly at their equators, protons approaching another proton’s poles along the spin axis meet the least charge resistance. At some point the approach will halt due to mutual and opposite proton charge emissions. It seems that it may or may not be necessary that some additional energy is needed to overcome the pole line’s minimum resistance, a ratcheting increased energy hurdle that needs to be cleared in order to get close enough to secure either the true parallel or anti-parallel atomic bond. In those bond positions there’s still sufficient proton emissions to prevent the protons from coming any closer together – equal to the bond gap distance between parallel or anti-parallel protons.
Now for a possible orthogonal bond. Imagine any type ep stack with either its low-cfz or electron occupied proton pole end approaching a nearby alpha T/B center from any direction within the alpha center equatorial plane. Traveling straight or end-over-end, the path between the two parallel, adjacent, vertical spin axis emission planes is narrow, but the incoming stack avoids the main emissions, encountering a minimum charge resistance where eventually a drifting approach will be halted. There needs to be enough energy to deliver the approaching stack’s lead proton (or other proton within the approaching stack) to end up directly between the T/B alpha’s center ep proton poles in one or both of its low-cfz’s, where it will feed its equatorial emisions into the alpha’s top and bottom protons, forming a stable ortho bond. Like splitting a target, there may have been some T/B alpha top and bottom ep bond gap distance increase resizing. It may be true that ortho bond gaps can be shorter than parallel or anti-parallel bond gaps. Or starting from a single orthogonal proton at an atom’s otherwise unoccupied (0,0,0) coordinate low-cfz center I can imagine adding one ep at a time to create either the +/- x axis (stacks 8 and 9) or +/- y axis (stacks 7 and 6) carousel arms.
Note that if the target stack was not a T/B type alpha center, the incoming ortho stack could cleave and break the target stack in two. The incoming stack captures and occupies the target stack’s single available internal low-cfz which was previously one of the bonds holding that stack together.
Having a z axis T/B alpha (slot1) and the makings for the first orthogonal (say) +/- x axis carousel legs’ center pinned to the z axis charge channel, how can a second orthogonal set, +/- y axis aligned carousel legs be added? That’s where the T/B alpha’s second low-cfz may come in handy. The second carousel’s +/- y axis oriented stack, can also fit into the T/B alpha’s vertically extended two low-cfz’s center either above or below the x oriented proton at the T/B alpha’s center in the same manner as the first – in the same z oriented minimum between the two vertically adjacent, proton equatorial emission planes, but at the same time approaching perpendicular (orthogonally) from the y direction between side adjacent +/- x plane oriented proton emissions. Which, together with adjacent +/- z plane emissions resembles a four sided minimum emission shaft leading to the center of the T/B alpha. Somehow, the two overlapping(?), vertically extendable low-cfz’s are sufficient to also hold stack 1 and both vertically adjacent +/- x and +/- y equatorial emissions oriented protons sharing slot1’s +/- z axis charge channel all together. It may be that the four carousel arms can, in effect, form like two six-stack T’s and two six-stack B’s with their own extended double low-cfz’s as well. Once formed those stacks may be locked into place by nearby proton emissions. Still thinking about it. And how it might change the previous 144 orthogonal bonds graphic.
As we know, this is, at best, educated guesswork. Please feel free to add, comment, and/or criticize. You’re also fairly good at motivation and inspiration. I'm still wondering about formulas that can should include these low-cfz's.
*
http://milesmathis.com/index.html
332. Diatomic Hydrogen. My new charge bonding explains this much better than electron sharing. Plus an analysis of spin isomers. 9pp. http://milesmathis.com/diatom.pdf
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
We might need to reread this one. Like what can "grow" or "rearrange" with more C.F. added? :
214. Splitting the Electron?
http://milesmathis.com/cu.pdf
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Recall that in my last post I was considering two orthogonal protons within the T/B alpha’s two low charge field zones (low-cf’s). Basically I was wondering how the carousel could be created.
Thanks Cr6, I enjoyed re-reading “Splitting the Electron? No, not really”, a couple of times; actually my book copy, the 15th paper in Miles’ “Navigating the Nucleus”. An excellent choice – it succeeded in providing me a few good zen whacks to my understanding. Even Miles’ atomic charge diagrams threw me into mental confusion - seemingly at odds with my more recent discreet ep particle diagrams.

When Miles says “It fits right in the middle there, doesn't it?” he’s referring how well his charge field copper atomic diagram fits the Wiki image. This image combines the two and adds Fe and Cu slotlayout diagrams for discussion.
Airman. All well and good. Don’t ask me to describe the Jahn-Teller effect with its reported evidence of proposed electron degenerate particles: spinon, orbiton and halon. I didn’t seek out the referenced paper nor the Nature article. As Miles so often and aptly explains, today’s mainstream physics theories are usually sheer nonsense. I’ve given up trying to understand most of them long ago. Miles’ charge field descriptions however are explained in true physics form and make plenty of sense.Miles wrote. Now, I ask you to compare my Copper nucleus to the Cu(OH2) diagram from Wiki. It fits right in the middle there, doesn't it? And I didn't draw this Copper model to solve this problem. If you study my models for other elements from my nuclear.pdf paper and other papers, you will see that my diagram of Copper is the result of simple construction rules I laid down there—before I ever began studying this Cu-O problem. Specifically, we fill the noble levels 1 and 2 first, then add the carousel level. Iron completes that level, and Copper is three protons into the fourth level. Notice that the nucleus in period 4 is basically ten protons tall and seven protons wide, there is more potential difference top to bottom than side to side. This will help us solve this problem in a straightforward way. Nothing in my model is determined by math or ad hoc theory. It is determined by logic and mechanics. It is determined by what is necessary to physically channel the charge field through the nucleus.
You will say, “That is charge, but what you are plugging into these positions is protons, not charge.” But all charged particles follow charge. …
That doesn’t mean I don’t have difficulty in understanding, I certainly do. I’m still trying to understand how Miles accounts for the difference between axial and carousel bond length as in the paper’s next few paragraphs.
Airman. I’m also in the middle of re-reading the two follow-on papers Miles identifies in the last paragraph, “More on the Orbiton a close reading of the Nature letter” http://milesmathis.com/orbiton.pdf and “The Charge Profile of Sr2CuO3” http://milesmathis.com/orbiton2.pdf.Miles wrote. This explains the longer axial bonds of Cu-O in a natural way. It isn't that the bonds are longer, it is that the nucleus of Copper is actually taller than it is wide. You will say, “That isn't borne out by the numbers, which are not in a 10 to 7 ratio. … .
The slotlayout diagrams shows that in our current atomic data, neither Iron nor Copper contain any hook position protons.
I admit it seems I’ve lost sight of the charge field source in my drifting, incoming orthogonal proton descriptions. In my defense, engineers usually think in terms of building things - as in this project, and the fact that Miles allows that the elements were likely formed under great charge pressures as might be present in solar cores where such directed proton path drifting may be possible.
Anyway, at that point my reading was interrupted. The two T/B alpha vertically adjacent internal orthogonal protons I’d imagined - actually ep’s with arbitrary proton spin, left or right, accompanied by electrons at one of the two proton pole positions, collapsed into a single 2 ep stack, orthogonal to and on opposite sides of the z axis charge channel. One of the two protons experienced had an unexpected 90 degree twist - easy enough in the presence of the main z axis charge channel, then the two ep protons configured into a valid ep stack, occupying the plus and minus orthogonal axis positions on opposite sides of the z-axis charge channel.
The low charge emission shaft leading into the T/B alpha is still in place. Two new orthogonal ep’s may fit into the T/B alpha’s now vacant double low-cfz’s center. When fully occupied, another 90 rotation of just one of them may result in the creation of the second z orthogonal axis protons with its two ep’s located in the new axis’s + and – y axis positions. Mutual repulsion with the first orthogonal axis causes both orthogonal axis’s (from the z axis) to become orthogonal from each other. The T/B alpha has been converted into a carousal hub with single ep’s in slots 6,7,8 and 9.
Note that with the addition of the third pair of parallel adjacent proton emission planes, two walls of proton emissions are now crossing the previously clear low charge emission shaft passing through the T/B alpha center making it much more difficult to deliver any more protons to the T/B alpha's center.
I don’t know about you, but I’m liking this idea that pairs of orthogonal ep’s from slot1's T/B alpha center can become either the x and y carousel legs.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Still thinking about how orthogonal protons can form a carousel. There’s also the quote from “Splitting the Electron? No, not really”, providing some building direction.
In “How to Build the Elements”,Miles wrote. Specifically, we fill the noble levels 1 and 2 first, then add the carousel level.
Changed the grey discs to blue to agree with the two ep stack convention. Neon’s carousel form contains alphas in slots 1,6,7,8,9. Neon’s axial form contains alphas in slots 1,2,3,4,5.Miles wrote. Actually, Neon can (or could) find great stability in one of two shapes, both of which have ten neutrons. To diagram this, I will simplify the alpha particle into a single disk.
The first atom meeting the condition of filled noble levels 1 and 2 and a carousel is Silicon, Si, atomic number 14. I also included a plot of S, including the spin axis between each alpha's proton pair for clarity.
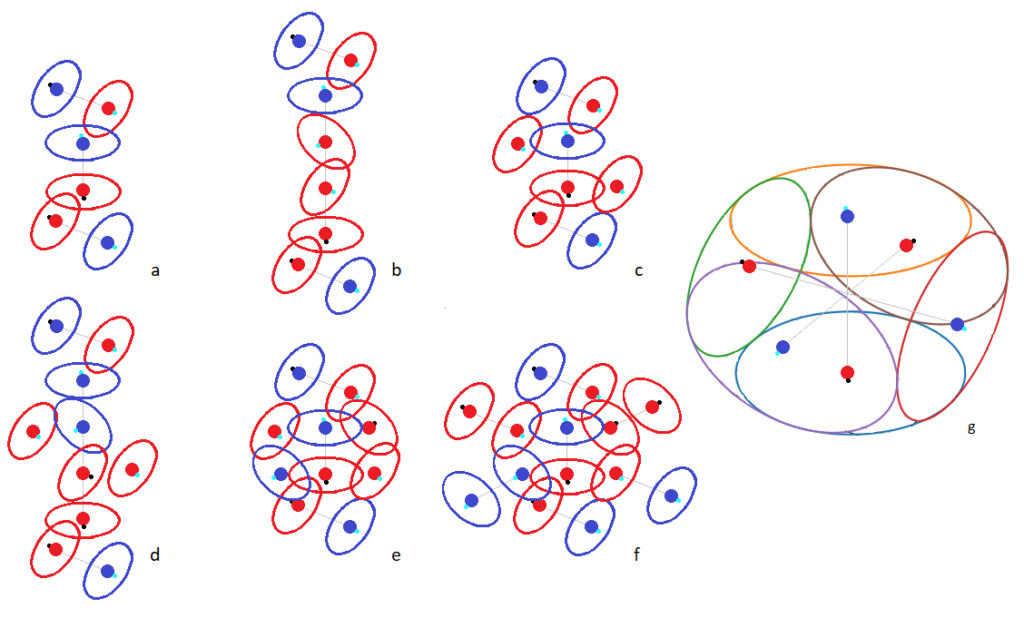
a – f. is intended to try to show how Silicon might be ‘grown’ starting from alpha pairs of ep’s in slots 1, 2 and 3 as I described last week. The usual form of Carbon Miles has shown contains two each alpha proton (ep) pairs in slot1 and single ep sets in slots 2 and 3. I suppose the Alpha filled slots 1, 2 and 3 containing six protons as shown in A might be an alternative, stable form of Carbon.
After having gone through the a-f exercise, I believe having the second noble level filled should increase the z axis charge channel bond tightness, which may be an important factor when building the carousel. In all honesty I became less enamored with the idea of adding individual orthogonal protons one at a time within slot1 with those ortho pairs then collapsing into either the +/- x or y legs as I went along.
g. This alternative view is useful and seems to suggest an an easier way to build a carousel - by directly adding three orthogonal alphas. Each proton in each x, y and z aligned alpha proton may be seen as occupying the centers of the opposite sides of six cubic faces. Unfortunately matplotlib has flattened the cube by half so it looks more like an ottoman. In any case, starting with the T/B slot1 z-aligned alpha, just slide a well positioned y-aligned alpha to straddle the z axis and then slide in an x-aligned alpha that also straddles the z axis. Each carousel leg is then a single ep. Single ep’s may then be added to the +/- x and y faces to form the Neon carousel.
Please pardon the delay this week. Plenty of unexpected real world tasks and less than efficient use of my project time – such as in re-re-reading the logic behind axial vs. carousel bonding lengths, and using matplotlib.
8/30/2012. Splitting the Electron? No, not really.
http://milesmathis.com/cu.pdf I have made a couple of corrections to this paper, to match it to findings in my two newer papers on the subject. My nuclear diagrams were a bit off, which threw off my math. I was able to correct the math on bond lengths, making it match data much better.
315. How to Build the Elements. http://milesmathis.com/nuclear.pdf
Explaining the periodic table, with nuclear diagrams. 71pp.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Cr6, please pardon this overdue progress update. Lost my mind in the meantime, taking a break from the subject whether I wanted to or not.
Last time I argued that the simplest way to build a carousel may be to add orthogonal alphas directly. But that violates the rule shown in my Thu, 26 Oct orthogonal bonding chart of 36 panels of four ortho bonds each – in which a proton must occupy the low charge pressure zone. Even a z-axis T/B alpha with two internal orthogonal protons collapsing into an x or y axis would require straddling the z axis and violating the occupied low-cfz rule.
Maybe somehow the dual internal opposite and overlapping low charge pressure zones within each orthogonal T/B alpha are needed for the proper balance and spacing of a carousel hub. This special set of double - 0 bond types are identified in their own chart above.
Does that make or break any charge field sense to you?
P.S. Added more double-0 combinations to the chart.
.
Last edited by LongtimeAirman on Sun Nov 19, 2023 10:16 pm; edited 1 time in total (Reason for editing : Added P.S.)
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
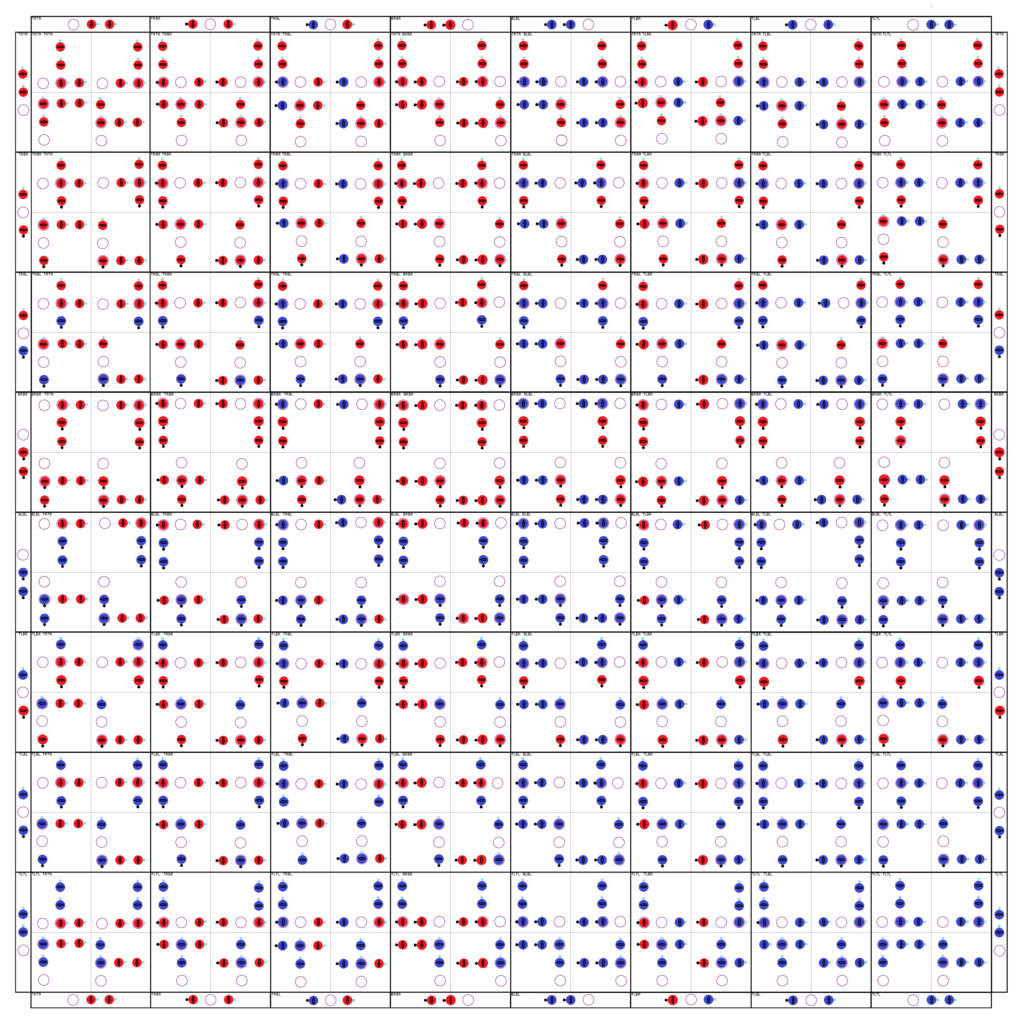
My current working assumption is that there are two different types of molecular bonds between two individual orthogonal 2-proton (ep) alpha stacks, ‘single’ or ‘double’.
All eight alphas (TRTR, TRBR, TRBL, BRBR, BLBL, TLBR, TLBL, TLTL) can form single low charge pressure zones (low-cfz) bonds. Single low-cfz orthogonal bond locations must be occupied by a single proton as shown in all the bonds above. Previously I’d posted an incomplete chart of 36 panels with 144 single bonds, here all possible single low-cfz bonds totals 8x8x4, or 256. They need to be verified somehow, some or all may or may not be valid.
Double bonds can presumably occur between orthogonal T/B alphas (TRBR, TRBL, TLBR, TLBL). Each T/B alpha contains two central overlapping low-cfz’s. In double low-cfz bonds a proton does not occupy the bonding location. The two T/B alphas in a double low-cfz fit together like a cross. The 16 orthogonal bond chart I posted last time (on 19 November) included all possible double type orthogonal bonds between two each two proton (ep) stacks. Double bonds may be needed to create a carousel.
There may also be other molecular ortho bonds not yet considered.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Slow progress this week, like dotting i's T or B and crossing t’s L or R.
The previous charts included all possible (?) ‘single’ and ‘double’ low charge pressure zone type bonds between a vertically oriented two e-p alpha stack, and a two e-p alpha stack rotated - orthogonally, 90 degrees clockwise. For comparison sake, today’s two charts also include ccw alpha rotations.
The above chart shows the 16 T/B alpha double low-cfz ortho bonds I’d posted previously. Here, four new columns have been added on the left; the total number of two e-p double low-cfz orthogonal (cw or ccw) bonds now total 32.

The second chart shows 12 of the possible 128 panels (64 cw and 64 ccw), each with four single bonds. 48 of the total 512 possible single low-cfz ortho bonds between vertical and any cw or ccw two e-p stack alphas.
Note that each of the two, cw and ccw horizontal rotations are 180 degree rotations from the other. Between the two horizontal rotations, both top/bottom electron positions and proton spin directions are switched.
A reminder that its not a simple task to determine whether any possible orthogonal bond is valid or not.
Cr6, Unless I’m mistaken - always a good chance, I believe the current ortho bond charts should satisfy your request for matrix type solutions. I've no idea how, but expressing this info as a formula should be possible.
If you still feel that way, please let me know exactly what you have in mind. I certainly don't know how you might script the sql ortho bonding logic, but I am at a point where I could create a new matplitlib plot for any of these e-p (2-6) alpha stack ortho bonds. Such plots might help in determining specific bond validities? They might also be a step toward actually plotting a molecule.Cr6 wrote. Nice graphic LTAM! Helped me see the alignments in a new perspective. I could work on adding a few via jupyter notebooks. May need a formula drop down widget that could try to show layout.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
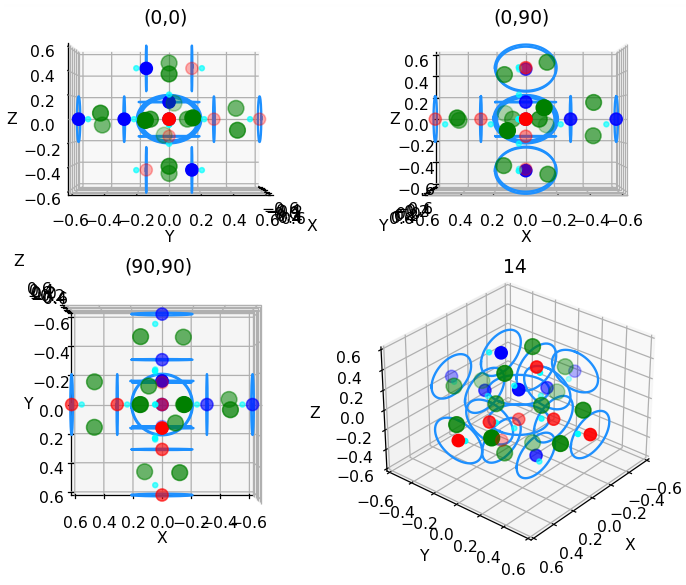
Silicon. Si, 14. The latest charge field atomic epn matplotlib plot, now with proton equatorial emission circles!
On 9 Nov23, while looking at growing a carousel, I included Si’s slotlayout diagram and previous epn plot. I’d also built a series of images of separate protons and circular plots - assembled by hand, adding up to Si, with a six-sided flattened ‘ottoman’ at its core.
My latest progress was to program the addition of circular plot equatorial emissions to all the protons, color coded to agree with each atom’s slotlayout diagram, as shown. The emission radius is easily adjustable, but if the circles get too large the image can easily become very confusing.
The z axis is still flatter than x or y but it looks good to me. Sorry it took so long, slow progress is better than no progress. I uploaded the changes to github.
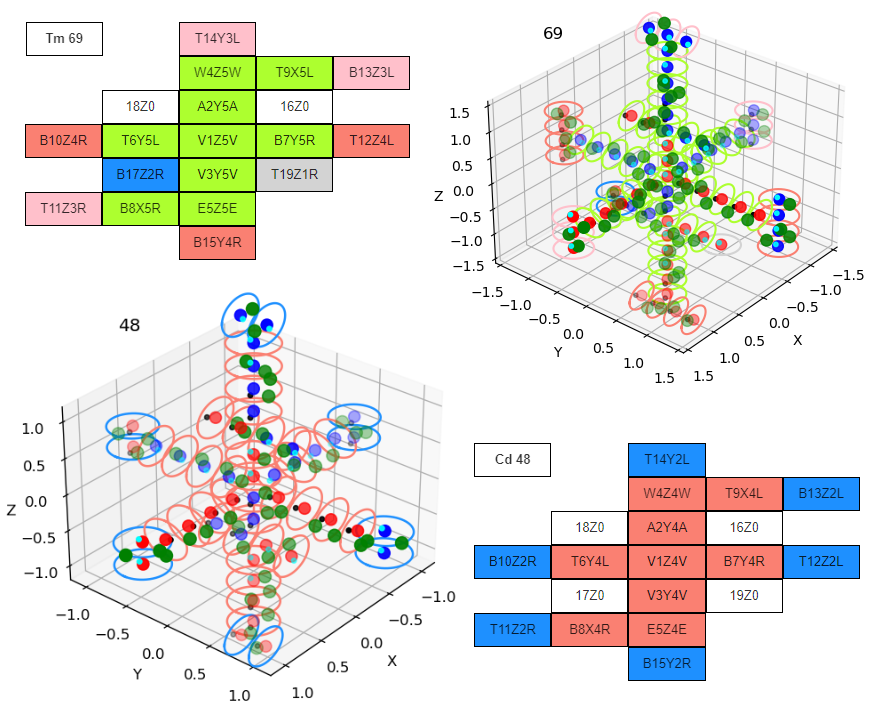
Thulium, Tm, 69 and Cadmium, Cd, 48.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
LongtimeAirman wrote:.
Silicon. Si, 14. The latest charge field atomic epn matplotlib plot, now with proton equatorial emission circles!
On 9 Nov23, while looking at growing a carousel, I included Si’s slotlayout diagram and previous epn plot. I’d also built a series of images of separate protons and circular plots - assembled by hand, adding up to Si, with a six-sided flattened ‘ottoman’ at its core.
My latest progress was to program the addition of circular plot equatorial emissions to all the protons, color coded to agree with each atom’s slotlayout diagram, as shown. The emission radius is easily adjustable, but if the circles get too large the image can easily become very confusing.
The z axis is still flatter than x or y but it looks good to me. Sorry it took so long, slow progress is better than no progress. I uploaded the changes to github.
Thulium, Tm, 69 and Cadmium, Cd, 48.
.
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
With respect to charge field atomic plots with proton equatorial emission circles,
Thanks Cr6. I am embarrassed it took me so long but I think it's a definite improvement.Cr6 wrote. Wow! Very, very cool LTAM!
With respect to an ortho bonding graphic, on
24 Oct 23 Cr6 wrote. I could work on adding a few via jupyter notebooks. May need a formula drop down widget that could try to show layout.
I’ve given your comment plenty of thought. For one thing, its unusual to hear you thinking about making notebook changes. If you want to do some scripting, feel free. I don’t want to step on your toes, I should try to assist you however you like rather than do it myself. Second it appears you are asking for: 1. a user interface widget containing drop down choices of proton stacks; and/or 2. a widget layout to display the selected bond; like the slotlayout diagram - tab enclosure widget. I’d immediately assumed the output would be another matplotlib plot – displaying the epn’s and bond between two proton stacks instead of any given atomic plot containing up to 19 occupied slots. Plus I’m not at all sure what constitutes a formula. I’m sure you understand my confusion. Please correct or direct me as you wish.
User interface epn stack selection. The user is expected to enter the following: 1. Two sets of electron, proton, neutron (epn) stack data; 2. The orientation of each stack, +/- x, y or z; and 3. The desired bond site.
Formula drop down widget. The user will be expected to select two different electron, proton neutron (epn) stacks.
Single list of 84 epn choices. Here’s a header row and a list of all the valid 1-6 epn stacks.
- Code:
t 1 2 3 4 5 6 bt b
TL b
t BR
TR b
t BL
TL TL b
t BR BR
TR TR b
t BL BL
TL bt BR
TR bt BL
TR bt BR
TL bt BL
TL TL TL b
t BR BR BR
TR TR TR b
t BL BL BL
TL bt BR BR
TR bt BL BL
TR bt BR BR
TL bt BL BL
TL TL bt BR
TR TR bt BL
TR TR bt BR
TL TL bt BL
TL TL TL TL b
t BR BR BR BR
TR TR TR TR b
t BL BL BL BL
TL bt BR BR BR
TR bt BL BL BL
TR bt BR BR BR
TL bt BL BL BL
TL TL bt BR BR
TR TR bt BL BL
TR TR bt BR BR
TL TL bt BL BL
TL TL TL bt BR
TR TR TR bt BL
TR TR TR bt BR
TL TL TL bt BL
TL TL TL TL TL b
t BR BR BR BR BR
TR TR TR TR TR b
t BL BL BL BL BL
TL bt BR BR BR BR
TR bt BL BL BL BL
TR bt BR BR BR BR
TL bt BL BL BL BL
TL TL bt BR BR BR
TR TR bt BL BL BL
TR TR bt BR BR BR
TL TL bt BL BL BL
TL TL TL bt BR BR
TR TR TR bt BL BL
TR TR TR bt BR BR
TL TL TL bt BL BL
TL TL TL TL bt BR
TR TR TR TR bt BL
TR TR TR TR bt BR
TL TL TL TL bt BL
TL TL TL TL TL TL b
t BR BR BR BR BR BR
TR TR TR TR TR TR b
t BL BL BL BL BL BL
TL bt BR BR BR BR BR
TR bt BL BL BL BL BL
TR bt BR BR BR BR BR
TL bt BL BL BL BL BL
TL TL bt BR BR BR BR
TR TR bt BL BL BL BL
TR TR bt BR BR BR BR
TL TL bt BL BL BL BL
TL TL TL bt BR BR BR
TR TR TR bt BL BL BL
TR TR TR bt BR BR BR
TL TL TL bt BL BL BL
TL TL TL TL bt BR BR
TR TR TR TR bt BL BL
TR TR TR TR bt BR BR
TL TL TL TL bt BL BL
TL TL TL TL TL bt BR
TR TR TR TR TR bt BL
TR TR TR TR TR bt BR
TL TL TL TL TL bt BL
A series of Drop down selections. Rather than selecting an epn directly from a list of 84 alternatives, we might also provide a series of drop downs. The number of user selections would depend on the number and type of epn tack desired.
1. Identify the number of epn’s (1-6) for a given stack. A drop down of 1-6.
2. Single epn. If a single epn (a stack of 1) is selected, that epn’s electron position and proton spin must also be selected. A drop down of: TR, BR, BL or TL.
3. Two-six epn stack. When a 2-6 epn stack is selected, The user must also identify the stack’s type. A dropdown of: TRTR, TRBR, TRBL, BRBR, BLBL, TLBR, TLBL, TLTL.
4a. Three-six epn stack with constant position and constant spin. Types: TRTR, BRBR, BLBL, TLTL. Each electron position and proton spin direction are the same. No further user input is needed. Any additional epn’s will also be the same as has already been identified.
4b. Three-six epn stack and TCount value. Three-six epn stacks of TB type: TRBR, TRBL, TLBR, TLBL involve a change of the electron position - from a T to B (top to bottom) proton poles between two adjacent protons within the vertically aligned 3-6 epn stack. A change in proton spin may or may not occur, the TCount value remains the same. The user must indicate how many T electron position epn’s the stack’s top contains. A drop down for the TCount value: (1-5).
5. Identify each epn stack’s orientation. +/- X, +/-Y or +/-Z. All the stacks within a given atom are defined according to +X, +Y and +Z. The bond plot should be able to determine the validity of a bond given any two of the 84 epn’s in any x, y or z orientation.
6. Identify the desired bonding location. Identify one or both of the desired bonding locations. Or it may be possible to show all possible bonds between the two selected stacks.
Do you want to add some script? Does the above and previous ortho bond posts help? I like the idea of some sort of bond widget and would end up trying to make one if you don’t. Please pardon me for being so pushy.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
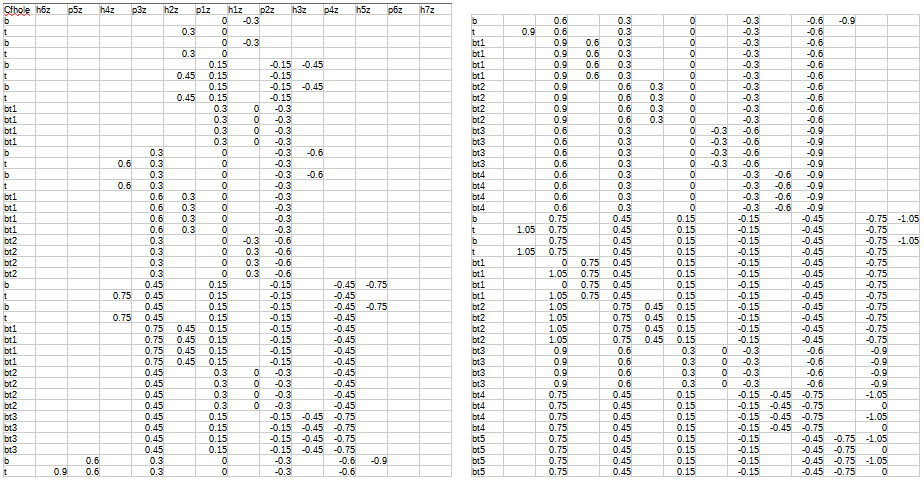
Project update. With the ‘bond widget’ in mind, I’m working on a small new database – EPNStacks.csv, independent of the atomic data, containing 84 rows, one for each of the allowed ep stack types. I’m trying to make the scheme as clear as possible. This image, with the first 42 rows on the left and second 43-84 rows on the right, shows the z coordinates for each ep stack’s protons and low charge pressure holes. The ep z centers (0,0,0) shown are currently at either P1 or (P2-P1)/2; although those two points are not always at each ep stack’s geometric z center, such as ep’s 77-84 with left (top) proton 5 at z=0.75 and right (bottom) side proton 6 at z=-1.05. That may or may not need correction. All x and y values equal 0. EPNStacks.csv also contains all (non-randomized, x or y, and z) neutron and electron coordinates - with respect to and offset from their associated slot z center and stack’s proton spin axis.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Thanks Cr6.
Latest progress. EPNStacks.csv. Thought more about it and decided to center each stack’s protons plus low-charge field hole z positions with that stack’s geometric center; as can be seen through all the additional zeros in the latest version image above.
Bond widget. Allowing the user to build up an epn stack with a logical series of dropdowns turned out to be a lot harder problem than I expected - it may not happen. I did the next best thing by adding another integer value to the 1-84 epn stack dropdown choices, (1-6), the number of protons in that stack, as can be seen in the image below. Reading long stack TBRL sequence is much easier with that proton value.
Added the code for a couple more dropdown widgets, selectOrient1 and selectOrient2.
Broke the 4 bond selection widgets into their own GUI.
Saved the changes to github.
Had to spend time reacquainting myself with pandas dataframe dictionary tuples.
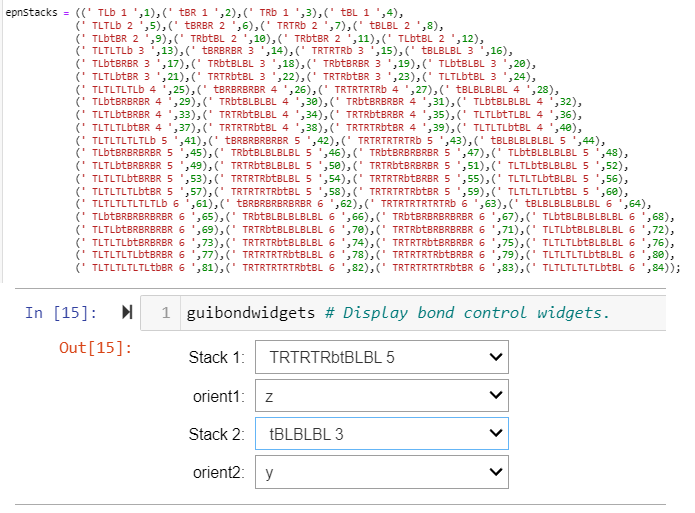
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
------------
DECEMBER 14, 2023
Editors' notes
Discovery of a new Hall effect: Does it violate Onsager's reciprocal theorem?
by University of Tsukuba
Correspondence among magnetotransport phenomena, centrosymmetric extended magnetic multipoles on pyrochlore lattice, and orbital shapes for magnetic (red and blue) and toroidal (green and magenta) charges. Isotropic anomalous Hall effect occurs on
a magnetic dipole order (ferromagnet).
b Extended magnetic toroidal quadrupole order allows anisotropic spin Hall effect but no electric charge Hall effect.
c Conical magnetic structure comprising the magnetic toroidal quadrupole and magnetic dipole shows anisotropic electric charge Hall effect.
Credit: Nature Communications (2023). DOI: 10.1038/s41467-023-43543-1
Researchers at the University of Tsukuba and NIMS have observed a novel Hall effect wherein the deflection direction of current varies according to the current flow direction. Onsager's reciprocal theorem, a fundamental theorem in materials science, asserts the impossibility of such a phenomenon. However, the researchers found that the observed phenomenon could be explained without contradicting the reciprocal theorem by assuming an unconventional magnetic arrangement.
The Hall effect, or anomalous Hall effect, occurs when electric current flows through a conductor or magnet in a magnetic field, generating voltage perpendicular to the electric and magnetic field directions. Onsager's reciprocal theorem states that the deflection direction of electrons remains constant, irrespective of the current's direction in the plane perpendicular to the magnetic field or magnetization.
In a new study, for the first time, researchers have observed an anisotropic anomalous Hall effect in a spinel oxide NiCo2O4 thin film with conical magnetic anisotropy; this characteristic depends on the current's direction. The findings are published in the journal Nature Communications.
To understand this phenomenon, researchers considered the symmetry of the experimentally observed anisotropic anomalous Hall effect from a phenomenological perspective. The findings indicated the involvement of a magnetic structure called a clustered magnetic toroidal quadrupole.
...
More information: Hiroki Koizumi et al, Quadrupole anomalous Hall effect in magnetically induced electron nematic state, Nature Communications (2023). DOI: 10.1038/s41467-023-43543-1
Journal information: Nature Communications
More at link: https://phys.org/news/2023-12-discovery-hall-effect-violate-onsager.html
---------
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

As per my current thinking, any given stack can contain 1-6 epn sets (electron proton and neutron), as well as a single low charge field pressure hole that may occupy one of seven possible stack locations, adjacent to or between protons; at either at the stack’s +z top (t), 1-5 middle positions (bt) or stack bottom (b) as highlighted by the seven colors above. Since last time, I added a new ‘Hloc’ column, identifying any given epn stack’s hole location by a label (h1z-h7z), indicating the correct hole column data for that stack, making the hole and proton locations easier to read and understand.
That image was quickly outdated by the addition of 26 new columns for all proton (1-6) and hole (1-7) x and y coordinates. In EPNStacks.csv these values were all zero, I didn’t think I needed them at first, but the notebook's rotation function will be used to convert the stack orientations to user selected orientations besides +z, such as -z, +/- x or +/-y, and using the rotation function makes it necessary for all the particles and holes to have all three x y and z coordinate values. So I decided it might be best to include the additional coordinate data fields in the EPNStacks.csv file.
Converted the reading EPNStacks.csv notebook cell code to read the added zero columns. As I may have mentioned recently, all numerical columns in the csv file contains floats, zeros or integers. Unassigned zero (0) x or y coordinates defaulted to longints. Converting the csv file coordinate zeros to 0.0 allowed pandas to correctly interpret those values as floats, so I was able to drop the no longer needed previous code converting those longint types to floats.
Added a new third value that appears on the epn drop down – 1-84, the number assigned to that epn configuration. The two digit decimal number is an easier way to identify the epn selected, compared to the 3-14 character epn TBRL and hole code.
Moved the “Bond widget” - and subsequent cell with initial draft code identifying particle locations for plotting, between the four atomic subplots and the Network Graphs sections of the notebook.
Slowly getting there.
Cr6 posted: Discovery of a new Hall effect: Does it violate Onsager's reciprocal theorem?
From https://phys.org/news/2023-12-discovery-hall-effect-violate-onsager.html
Quoting the story.
AndResearchers at the University of Tsukuba and NIMS have observed a novel Hall effect wherein the deflection direction of current varies according to the current flow direction. Onsager's reciprocal theorem, a fundamental theorem in materials science, asserts the impossibility of such a phenomenon. However, the researchers found that the observed phenomenon could be explained without contradicting the reciprocal theorem by assuming an unconventional magnetic arrangement.
Correspondence among magnetotransport phenomena, centrosymmetric extended magneticmultipoles on pyrochlore lattice, and orbital shapes for magnetic (red and blue) and toroidal (green and magenta) charges.
Thanks Cr6. I certainly could not understand any of the techno-jargon or details, but realized that the complex magnetic arrangement that the researchers referred to are likely due to charge field atomic slot layouts of atomic stacks. For a better understanding and possible future study, I was able to download the source link included in the Phy.org story, https://dx.doi.org/10.1038/s41467-023-43543-1 Nature Communications article “Quadrupole anomalous Hall effect in magnetically induced electron nematic state” which includes much more information and plenty of interesting diagrams.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Was trying to do this with orientdb but it tended to run out of memory on my laptop.
Apache Age:
https://github.com/apache/age
The file imports are really straight forward:
https://age.apache.org/age-manual/master/intro/agload.html#importing-graph-from-files
Python driver for Age:
https://github.com/apache/age/tree/master/drivers/python
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Brief project update. Corrected about 8 separate hole z coordinate decimal position typos that appeared in the highlighted image I posted on Tuesday, such as in changing 1.5 to 0.15, or changing -4.5 to -0.45.
Also changed the 'Hloc' column data values from: 'h1z', ..., 'h7z'; to: 'h1', ..., 'h7' - seemed like the right thing to do based on some principle or other. That made finding the actual columns containing the hole x,y,z coordinates a bit of a challenge. I answered that with a generous amount of code lines that seemed to do the job.
- Code:
"""
Bonding widget output under construction
A Bonding widget will allow closer study to help
determine the validity of bonds between adjacent
orthogonal epn stacks.
An epn (electrons, protons and neutrons) stack
consists of 1-6 epn sets. Each stack includes a
low charge field pressure zone (low-cfz) 'hole'
at either the stack's top, bottom, or somewhere
in the middle. I'll assume bonding between two
stacks should occur at one or both stack holes.
Find the x y and z coordinate locations for all the
protons, electrons, neutrons and hole for any given epn
set, given different stack orientations and positions.
"""
# These 12 lists includes the header 'loc's for all epn
# particles and holes' x y z coordinates.
# This series of 12 lists point to the columns
# containing up to 18 (x,y,z) particle coordinates
# and 7 (x,y,z) low-cfz hole coordinates.
eleXColList = ['e1x', 'e2x', 'e3x', 'e4x', 'e5x', 'e6x']
eleYColList = ['e1y', 'e2y', 'e3y', 'e4y', 'e5y', 'e6y']
eleZColList = ['e1z', 'e2z', 'e3z', 'e4z', 'e5z', 'e6z']
proXColList = ['p1x', 'p2x', 'p3x', 'p4x', 'p5x', 'p6x']
proYColList = ['p1y', 'p2y', 'p3y', 'p4y', 'p5y', 'p6y']
proZColList = ['p1z', 'p2z', 'p3z', 'p4z', 'p5z', 'p6z']
neuXColList = ['n1x', 'n2x', 'n3x', 'n4x', 'n5x', 'n6x']
neuYColList = ['n1y', 'n2y', 'n3y', 'n4y', 'n5y', 'n6y']
neuZColList = ['n1z', 'n2z', 'n3z', 'n4z', 'n5z', 'n6z']
#lowXColList = ['h1x', 'h2x', 'h3x', 'h4x', 'h5x', 'h6x', 'h7x']
#lowYColList = ['h1y', 'h2y', 'h3y', 'h4y', 'h5y', 'h6y', 'h7y']
#lowZColList = ['h1z', 'h2z', 'h3z', 'h4z', 'h5z', 'h6z', 'h7z']
# Left spinning Protons.
pLXs = []
pLYs = []
pLZs = []
# Right spinning Protons.
pRXs = []
pRYs = []
pRZs = []
# Top proton pole Electrons.
eTXs = []
eTYs = []
eTZs = []
# Bottom proton pole Electrons.
eBXs = []
eBYs = []
eBZs = []
# Neutrons.
nXs = []
nYs = []
nZs = []
# Holes.
hXs = []
hYs = []
hZs = []
pd.options.display.max_columns = None
pd.options.display.max_rows = None
# Copy the two selected epn rows from epnStacks to
# create single row dataframes epn1_df and epn2_df.
epn1_df = dfepn.loc[numberepn1-1, 'StackNumber':'e6z']
epn2_df = dfepn.loc[numberepn2-1, 'StackNumber':'e6z']
# Create a variable for the total number of protons
# in each stack.
epn1protons = epn1_df.loc['Protons']
epn2protons = epn2_df.loc['Protons']
#print('epn1_df',epn1_df,'epn2_df',epn2_df)
#print(epn1_df[:]) # Same output
# These hole variables and the code to select
# the correct (h1-h7) x,y,z coordinate column
# headers the data is in is cumbersome, and it
# needs to be done twice(!), yet it appears to
# work. There must be an easier way, i.e. function.
hole1 = epn1_df.loc['Hloc']
hole1X, hole1Y, hole1Z = 'holeXW', 'holeXW', 'holeXW'
if hole1 == 'h1':
hole1X, hole1Y, hole1Z = 'h1x', 'h1y', 'h1z'
if hole1 == 'h2':
hole1X, hole1Y, hole1Z = 'h2x', 'h2y', 'h2z'
if hole1 == 'h3':
hole1X, hole1Y, hole1Z = 'h3x', 'h3y', 'h3z'
if hole1 == 'h4':
hole1X, hole1Y, hole1Z = 'h4x', 'h4y', 'h4z'
if hole1 == 'h5':
hole1X, hole1Y, hole1Z = 'h1x', 'h1y', 'h1z'
if hole1 == 'h6':
hole1X, hole1Y, hole1Z = 'h6x', 'h6y', 'h6z'
if hole1 == 'h7':
hole1X, hole1Y, hole1Z = 'h7x', 'h7y', 'h7z'
hole2 = epn2_df.loc['Hloc']
hole2X, hole2Y, hole2Z = 'holeXW', 'holeXW', 'holeXW'
if hole2 == 'h1':
hole2X, hole2Y, hole2Z = 'h1x', 'h1y', 'h1z'
if hole2 == 'h2':
hole2X, hole2Y, hole2Z = 'h2x', 'h2y', 'h2z'
if hole2 == 'h3':
hole2X, hole2Y, hole2Z = 'h3x', 'h3y', 'h3z'
if hole2 == 'h4':
hole2X, hole2Y, hole2Z = 'h4x', 'h4y', 'h4z'
if hole2 == 'h5':
hole2X, hole2Y, hole2Z = 'h1x', 'h1y', 'h1z'
if hole2 == 'h6':
hole2X, hole2Y, hole2Z = 'h6x', 'h6y', 'h6z'
if hole2 == 'h7':
hole2X, hole2Y, hole2Z = 'h7x', 'h7y', 'h7z'
# All coordinates output below are taken directly from
# the two epn stack rows - no translations or rotations.
print('StackNumber = ', numberepn1 )
for i in range(epn1protons):
print('proton ',i+1,' x,y,z = ', epn1_df.loc[proXColList[i]],
epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]])
print('neutron ',i+1,' x,y,z = ', epn1_df.loc[neuXColList[i]],
epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]])
print('electron ',i+1,' x,y,z = ', epn1_df.loc[eleXColList[i]],
epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]])
print('hole x,y,z = ', epn1_df.loc[hole1X], epn1_df.loc[hole1Y],
epn1_df.loc[hole1Z], epn1_df.loc['Hloc'])
print('StackNumber = ', numberepn2 )
for j in range(epn2protons):
print('proton ',j+1,' x,y,z = ', epn2_df.loc[proXColList[j]],
epn2_df.loc[proYColList[j]], epn2_df.loc[proZColList[j]])
print('neutron ',j+1,' x,y,z = ', epn2_df.loc[neuXColList[j]],
epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]])
print('electron ',j+1,' x,y,z = ', epn2_df.loc[eleXColList[j]],
epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]])
print('hole x,y,z = ', epn2_df.loc[hole2X], epn2_df.loc[hole2Y],
epn2_df.loc[hole2Z], epn2_df.loc['Hloc'])
"""
Output:
StackNumber = 15
proton 1 x,y,z = 0 0 0.15
neutron 1 x,y,z = 0.15 0 -0.15
electron 1 x,y,z = 0.045 0 0.0648
proton 2 x,y,z = 0 0 -0.15
neutron 2 x,y,z = 0.15 0 -0.15
electron 2 x,y,z = 0.045 0 0.0648
proton 3 x,y,z = 0 0 0.45
neutron 3 x,y,z = 0.15 0 -0.15
electron 3 x,y,z = 0.045 0 0.0648
hole x,y,z = 0 0 -0.45 h3
StackNumber = 16
proton 1 x,y,z = 0 0 -0.15
neutron 1 x,y,z = 0.15 0 0.15
electron 1 x,y,z = 0.045 0 -0.0648
proton 2 x,y,z = 0 0 -0.45
neutron 2 x,y,z = 0.15 0 0.15
electron 2 x,y,z = 0.045 0 -0.0648
proton 3 x,y,z = 0 0 0.15
neutron 3 x,y,z = 0.15 0 0.15
electron 3 x,y,z = 0.045 0 -0.0648
hole x,y,z = 0 0 0.45 h4
"""
# Sort the epn particles, starting with protons -
# as with 'L' or 'R' spins. Electron positions
# near 'T' or 'B' proton pole.
# Similar to reading atoms from the atomic data.
#print("orient1 is ",orient1," and epn1_df.loc['SlotOrien'] = ",epn1_df.loc['SlotOrien'])
#print("orient2 is ",orient2," and epn2_df.loc['SlotOrien'] = ",epn2_df.loc['SlotOrien'])
"""
Plot z aligned epn' stacks before doing any rotations.
""";
# Rotate the two stacks to their selected orientations.
# In EPNStacks, proton and hole coordinates are listed in
# a +z epn alignment, for which all x and y values are 0.
# Z aligned electrons and neutrons however, do have z
# and (x or y) values for starters.
# Rotation function is copied here for
# convenient review,
#Rotates and returns a tuple (x, y, z) of the
#three x, y, z positions and spin axis angle entered.
#ax, ay or az are angles in radians about the three
#x y and z cubic spin axes through (0,0,0).
#def rotateAbtX(x, y, z, ax):
# rotatedX = x
# rotatedY = (y * cos(ax)) - (z * sin(ax))
# rotatedZ = (y * sin(ax)) + (z * cos(ax))
# x, y, z = rotatedX, rotatedY, rotatedZ
# return(rotatedX, rotatedY, rotatedZ )
#if orient1 == -z :
# Rotate pi rads about x or y or just flip the z coordinates.
# Two ways to do so, x or y are arbitrary and equally likely.
# for i in range(epn1protons):
# epn1PValues[i] = -1 * epn1PValues[i]epn1_df.
#if orient1 == x :
# Rotate +z pi/2 rads about y
# for i in range(epn1protons):
# epn1PValues[i] = -1 * epn1PValues[i]
# ...
My next task will be to sort the particles, identifying L or R protons and T or B electrons - in preparation for plotting. Simple single epn plots first, followed by two epn plot including translation and rotations. Its not difficult but I expect to be otherwise busy here over the next few days.
Hope everyone has a safe and happy Holiday and New Year.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
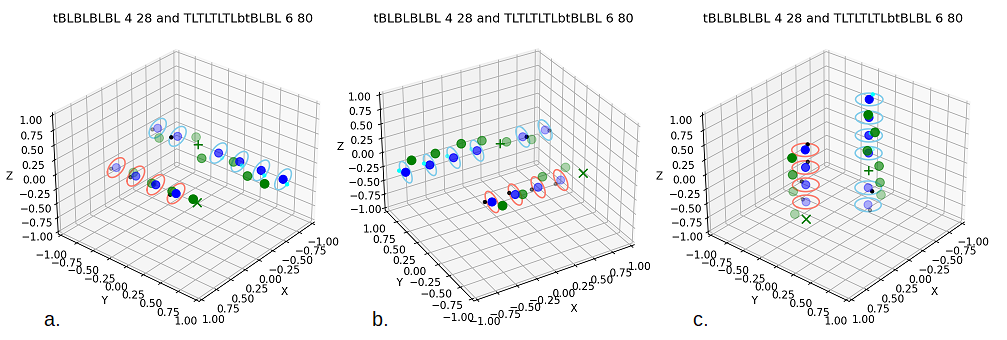
Bonding widget progress update. Two selected epn's (identified in the image's three titles), including electrons (B or T), neutrons, protons (L or R), proton emissions and holes are plotted in a demo sampler of possible +/-x, +/-y and +/-z orientations at temporary 'offset' coordinates and parallel/antiparallel orientations. a. +y, +y. b. +x, -x. c. -z, +z. There’s no reason why those orientations need be limited to just 90 degree orthogonals. The stack end with the cyan electron started at +z, or the black electron end started at -z. Offset1X, offset1Y, offset1Z = 0.25 -0.25 -0.25 and offset2X, offset2Y, offset2Z = -0.25 0.25 0.25. All epn’s have the same default (0,0,0) center and they might intersect and overlap to some extent if they were not offset. Those offsets need to be changed to each stack’s possible bonding site coordinates, the epn stack’s hole. Epn1’s hole is indicated by a green ‘X’, epn2’s hole by a green ‘+’. I don’t know how to determine what sort of logic might apply to determining valid stacks. The neutrons and electrons have been given random proton pole orbital positions. The plot title is not programmed or automated – but I think I see how I might be able to do that. I’d like to be able to change distances at will, or to turn things on or off. Please feel free to make any suggestions.
I can easily imagine replacing each and every stack (1-19) in any given atom by one (or be selected from a set of several possible) of the bonding widget’s 84 stacks. Does epnStacks.csv or this bonding widget make the search for each and every bond in the universe or your sql script any easier?
Things have gone smoothly enough, most of my problems stem from my own shortcomings and frequent typos. Here’s the Bonding widget cell’s code. I’m sure the 330 lines can be shortened, since it does everything twice - once for each epn stack. Replacing two occurrences with single functions would certainly be an improvement.
- Code:
"""
Bonding widget output under construction.
A Bonding widget will allow closer study to help
determine the validity of bonds between adjacent
orthogonal epn stacks.
An epn (electrons, protons and neutrons) stack
consists of 1-6 epn sets. Each stack includes a
low charge field pressure zone (low-cfz) 'hole'
at either the stack's top, bottom, or somewhere
in the middle. I'll assume bonding between two
stacks should occur at one or both stack holes.
Build selected epns.
"""
# These 12 lists includes the header 'loc's for all epn
# particles and holes' x y z coordinates.
# This series of 12 lists point to the columns
# containing up to 18 (x,y,z) particle coordinates
# and 7 (x,y,z) low-cfz hole coordinates.
# Added a proton spin col with values 'ps1',...,'ps6
# containing L or R proton spin and another row for
# each electron's T or B value, 'ep1',...,'ep6,
eleXColList = ['e1x', 'e2x', 'e3x', 'e4x', 'e5x', 'e6x']
eleYColList = ['e1y', 'e2y', 'e3y', 'e4y', 'e5y', 'e6y']
eleZColList = ['e1z', 'e2z', 'e3z', 'e4z', 'e5z', 'e6z']
elePoColList = ['ep1', 'ep2', 'ep3', 'ep4', 'ep5', 'ep6']
proXColList = ['p1x', 'p2x', 'p3x', 'p4x', 'p5x', 'p6x']
proYColList = ['p1y', 'p2y', 'p3y', 'p4y', 'p5y', 'p6y']
proZColList = ['p1z', 'p2z', 'p3z', 'p4z', 'p5z', 'p6z']
proSpColList = ['ps1', 'ps2', 'ps3', 'ps4', 'ps5', 'ps6']
neuXColList = ['n1x', 'n2x', 'n3x', 'n4x', 'n5x', 'n6x']
neuYColList = ['n1y', 'n2y', 'n3y', 'n4y', 'n5y', 'n6y']
neuZColList = ['n1z', 'n2z', 'n3z', 'n4z', 'n5z', 'n6z']
lowXColList = ['h1x', 'h2x', 'h3x', 'h4x', 'h5x', 'h6x', 'h7x']
lowYColList = ['h1y', 'h2y', 'h3y', 'h4y', 'h5y', 'h6y', 'h7y']
lowZColList = ['h1z', 'h2z', 'h3z', 'h4z', 'h5z', 'h6z', 'h7z']
# Initializing all the particle position lists.
# All protons in each epn stack
epn1AllpXs, epn1AllpYs, epn1AllpZs = [], [], []
epn2AllpXs, epn2AllpYs, epn2AllpZs = [], [], []
# Left spinning Protons.
epn1pLXs, epn1pLYs, epn1pLZs = [], [], []
epn2pLXs, epn2pLYs, epn2pLZs = [], [], []
# Right spinning Protons.
epn1pRXs, epn1pRYs, epn1pRZs = [], [], []
epn2pRXs, epn2pRYs, epn2pRZs = [], [], []
# All electrons
epn1AlleXs, epn1AlleYs, epn1AlleZs = [], [], []
epn2AlleXs, epn2AlleYs, epn2AlleZs = [], [], []
# Top proton pole Electrons.
epn1eTXs,epn1eTYs,epn1eTZs = [], [], []
epn2eTXs,epn2eTYs,epn2eTZs = [], [], []
# Bottom proton pole Electrons.
epn1eBXs,epn1eBYs,epn1eBZs = [], [], []
epn2eBXs,epn2eBYs,epn2eBZs = [], [], []
# Neutrons.
epn1nXs, epn1nYs, epn1nZs = [], [], []
epn2nXs, epn2nYs, epn2nZs = [], [], []
# Holes.
epn1hXs, epn1hYs, epn1hZs = [], [], []
epn2hXs, epn2hYs, epn2hZs = [], [], []
#pd.options.display.max_columns = None
#pd.options.display.max_rows = None
# Copy the two selected epn rows from epnStacks to
# create single row dataframes epn1_df and epn2_df.
epn1_df = dfepn.loc[numberepn1-1, 'StackNumber':'e6z']
epn2_df = dfepn.loc[numberepn2-1, 'StackNumber':'e6z']
# Create a variable for the total number of protons
# in each stack.
epn1protons = epn1_df.loc['Protons']
epn2protons = epn2_df.loc['Protons']
#print('epn1_df',epn1_df,'epn2_df',epn2_df)
#print(epn1_df[:]) # Same output
# These hole variables and the code to select
# the correct (h1-h7) x,y,z coordinate column
# headers the data is in is cumbersome, and it
# needs to be done twice(!), yet it appears to
# work. There must be an easier way, i.e. function.
hole1 = epn1_df.loc['Hloc']
hole1X, hole1Y, hole1Z = 'hole1XW', 'hole1YW', 'hole1ZW'
if hole1 == 'h1':
hole1X, hole1Y, hole1Z = 'h1x', 'h1y', 'h1z'
if hole1 == 'h2':
hole1X, hole1Y, hole1Z = 'h2x', 'h2y', 'h2z'
if hole1 == 'h3':
hole1X, hole1Y, hole1Z = 'h3x', 'h3y', 'h3z'
if hole1 == 'h4':
hole1X, hole1Y, hole1Z = 'h4x', 'h4y', 'h4z'
if hole1 == 'h5':
hole1X, hole1Y, hole1Z = 'h5x', 'h5y', 'h5z'
if hole1 == 'h6':
hole1X, hole1Y, hole1Z = 'h6x', 'h6y', 'h6z'
if hole1 == 'h7':
hole1X, hole1Y, hole1Z = 'h7x', 'h7y', 'h7z'
#print('hole x,y,z = ', epn1_df.loc[hole1X], epn1_df.loc[hole1Y],
# epn1_df.loc[hole1Z], epn1_df.loc['Hloc'])
hole2 = epn2_df.loc['Hloc']
hole2X, hole2Y, hole2Z = 'hole2XW', 'hole2YW', 'hole2ZW'
if hole2 == 'h1':
hole2X, hole2Y, hole2Z = 'h1x', 'h1y', 'h1z'
if hole2 == 'h2':
hole2X, hole2Y, hole2Z = 'h2x', 'h2y', 'h2z'
if hole2 == 'h3':
hole2X, hole2Y, hole2Z = 'h3x', 'h3y', 'h3z'
if hole2 == 'h4':
hole2X, hole2Y, hole2Z = 'h4x', 'h4y', 'h4z'
if hole2 == 'h5':
hole2X, hole2Y, hole2Z = 'h5x', 'h5y', 'h5z'
if hole2 == 'h6':
hole2X, hole2Y, hole2Z = 'h6x', 'h6y', 'h6z'
if hole2 == 'h7':
hole2X, hole2Y, hole2Z = 'h7x', 'h7y', 'h7z'
# Rotate the hole's initial +z orientation to the selected orientation.
if orient1 == '-z':
epn1_df.loc[hole1X], epn1_df.loc[hole1Y], epn1_df.loc[hole1Z] = rotateAbtX(epn1_df.loc[hole1X], epn1_df.loc[hole1Y], epn1_df.loc[hole1Z], PI_PHI)
elif orient1 == 'x':
epn1_df.loc[hole1X], epn1_df.loc[hole1Y], epn1_df.loc[hole1Z] = rotateAbtY(epn1_df.loc[hole1X], epn1_df.loc[hole1Y], epn1_df.loc[hole1Z], PI_PHI/2)
elif orient1 == '-x':
epn1_df.loc[hole1X], epn1_df.loc[hole1Y], epn1_df.loc[hole1Z] = rotateAbtY(epn1_df.loc[hole1X], epn1_df.loc[hole1Y], epn1_df.loc[hole1Z], -PI_PHI/2)
elif orient1 == 'y':
epn1_df.loc[hole1X], epn1_df.loc[hole1Y], epn1_df.loc[hole1Z] = rotateAbtX(epn1_df.loc[hole1X], epn1_df.loc[hole1Y], epn1_df.loc[hole1Z], -PI_PHI/2)
elif orient1 == '-y':
epn1_df.loc[hole1X], epn1_df.loc[hole1Y], epn1_df.loc[hole1Z] = rotateAbtX(epn1_df.loc[hole1X], epn1_df.loc[hole1Y], epn1_df.loc[hole1Z], PI_PHI/2)
epn1_df.loc[hole1X], epn1_df.loc[hole1Y], epn1_df.loc[hole1Z] = round(epn1_df.loc[hole1X],2), round(epn1_df.loc[hole1Y],2), round(epn1_df.loc[hole1Z],2)
if orient2 == '-z':
epn2_df.loc[hole2X], epn2_df.loc[hole2Y], epn2_df.loc[hole2Z] = rotateAbtX(epn2_df.loc[hole2X], epn2_df.loc[hole2Y], epn2_df.loc[hole2Z], PI_PHI)
elif orient2 == 'x':
epn2_df.loc[hole2X], epn2_df.loc[hole2Y], epn2_df.loc[hole2Z] = rotateAbtY(epn2_df.loc[hole2X], epn2_df.loc[hole2Y], epn2_df.loc[hole2Z], PI_PHI/2)
elif orient2 == '-x':
epn2_df.loc[hole2X], epn2_df.loc[hole2Y], epn2_df.loc[hole2Z] = rotateAbtY(epn2_df.loc[hole2X], epn2_df.loc[hole2Y], epn2_df.loc[hole2Z], -PI_PHI/2)
elif orient2 == 'y':
epn2_df.loc[hole2X], epn2_df.loc[hole2Y], epn2_df.loc[hole2Z] = rotateAbtX(epn2_df.loc[hole2X], epn2_df.loc[hole2Y], epn2_df.loc[hole2Z], -PI_PHI/2)
elif orient2 == '-y':
epn2_df.loc[hole2X], epn2_df.loc[hole2Y], epn2_df.loc[hole2Z] = rotateAbtX(epn2_df.loc[hole2X], epn2_df.loc[hole2Y], epn2_df.loc[hole2Z], PI_PHI/2)
epn2_df.loc[hole2X], epn2_df.loc[hole2Y], epn2_df.loc[hole2Z] = round(epn2_df.loc[hole2X],2), round(epn2_df.loc[hole2Y],2), round(epn2_df.loc[hole2Z],2)
# If the holes are reoriented, orient the remaining
# stack particles to the selected direction.
# Moves the epn1 stack from its default (0,0,0) positions.
offset1X, offset1Y, offset1Z = 0.25, -0.25, -0.25
for i in range (epn1protons):
# Rotate the stack, particle by particle, to the deired spin
# axis direction. epn1 dataframe's 'SlotOrien' value is updated.
# Another possibility
#epn1px = epn1_df.loc[proXColList[i]]
#epn1py = epn1_df.loc[proYColList[i]]
#epn1pz = epn1_df.loc[proZColList[i]]
#epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]] = epn1px,epn1py,epn1pz
# Randomize the dataframe's fixed
# electron and neutron coordinates.
aze1 = random()*PI_PHI
azn1 = random()*PI_PHI
epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]] = rotateAbtZ(epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]], aze1)
epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]] = rotateAbtZ(epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]], azn1)
if orient1 == '-z':
epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]] = rotateAbtX(epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]], PI_PHI)
epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]] = rotateAbtX(epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]], PI_PHI)
epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]] = rotateAbtX(epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]], PI_PHI)
epn1_df.loc['SlotOrien'] = '-Z'
elif orient1 == 'x':
epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]] = rotateAbtY(epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]], PI_PHI/2)
epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]] = rotateAbtY(epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]], PI_PHI/2)
epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]] = rotateAbtY(epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]], PI_PHI/2)
epn1_df.loc['SlotOrien'] = 'X'
elif orient1 == '-x':
epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]] = rotateAbtY(epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]], -PI_PHI/2)
epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]] = rotateAbtY(epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]], -PI_PHI/2)
epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]] = rotateAbtY(epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]], -PI_PHI/2)
epn1_df.loc['SlotOrien'] = '-X'
elif orient1 == 'y':
epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]] = rotateAbtX(epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]], -PI_PHI/2)
epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]] = rotateAbtX(epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]], -PI_PHI/2)
epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]] = rotateAbtX(epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]], -PI_PHI/2)
epn1_df.loc['SlotOrien'] = 'Y'
elif orient1 == '-y':
epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]] = rotateAbtX(epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]], PI_PHI/2)
epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]] = rotateAbtX(epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]], PI_PHI/2)
epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]] = rotateAbtX(epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]], PI_PHI/2)
epn1_df.loc['SlotOrien'] = '-Y'
else :
epn1_df.loc['SlotOrien'] = 'Z'
# Append the epn's particle type and coordinates to
# their respective five categorical lists for plotting.
# All protons
epn1AllpXs.append(epn1_df.loc[proXColList[i]] + offset1X)
epn1AllpYs.append(epn1_df.loc[proYColList[i]] + offset1Y)
epn1AllpZs.append(epn1_df.loc[proZColList[i]] + offset1Z)
# Protons - Left or Right
if epn1_df.loc[proSpColList[i]] == 'L':
epn1pLXs.append(epn1_df.loc[proXColList[i]] + offset1X)
epn1pLYs.append(epn1_df.loc[proYColList[i]] + offset1Y)
epn1pLZs.append(epn1_df.loc[proZColList[i]] + offset1Z)
else :
epn1pRXs.append(epn1_df.loc[proXColList[i]] + offset1X)
epn1pRYs.append(epn1_df.loc[proYColList[i]] + offset1Y)
epn1pRZs.append(epn1_df.loc[proZColList[i]] + offset1Z)
# All Electrons
epn1AlleXs.append(epn1_df.loc[eleXColList[i]]+epn1_df.loc[proXColList[i]] + offset1X)
epn1AlleYs.append(epn1_df.loc[eleYColList[i]]+epn1_df.loc[proYColList[i]] + offset1Y)
epn1AlleZs.append(epn1_df.loc[eleZColList[i]]+epn1_df.loc[proZColList[i]] + offset1Z)
# Electrons - Top or Bottom
if epn1_df.loc[elePoColList[i]] == 'T':
epn1eTXs.append(epn1_df.loc[eleXColList[i]]+epn1_df.loc[proXColList[i]] + offset1X)
epn1eTYs.append(epn1_df.loc[eleYColList[i]]+epn1_df.loc[proYColList[i]] + offset1Y)
epn1eTZs.append(epn1_df.loc[eleZColList[i]]+epn1_df.loc[proZColList[i]] + offset1Z)
else :
epn1eBXs.append(epn1_df.loc[eleXColList[i]]+epn1_df.loc[proXColList[i]] + offset1X)
epn1eBYs.append(epn1_df.loc[eleYColList[i]]+epn1_df.loc[proYColList[i]] + offset1Y)
epn1eBZs.append(epn1_df.loc[eleZColList[i]]+epn1_df.loc[proZColList[i]] + offset1Z)
epn1nXs.append(epn1_df.loc[neuXColList[i]]+epn1_df.loc[proXColList[i]] + offset1X)
epn1nYs.append(epn1_df.loc[neuYColList[i]]+epn1_df.loc[proYColList[i]] + offset1Y)
epn1nZs.append(epn1_df.loc[neuZColList[i]]+epn1_df.loc[proZColList[i]] + offset1Z)
if orient1 == '-z':
epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]] = rotateAbtX(epn1_df.loc[proXColList[i]], epn1_df.loc[proYColList[i]], epn1_df.loc[proZColList[i]], PI_PHI)
epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]] = rotateAbtX(epn1_df.loc[eleXColList[i]], epn1_df.loc[eleYColList[i]], epn1_df.loc[eleZColList[i]], PI_PHI)
epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]] = rotateAbtX(epn1_df.loc[neuXColList[i]], epn1_df.loc[neuYColList[i]], epn1_df.loc[neuZColList[i]], PI_PHI)
epn1_df.loc['SlotOrien'] = '-Z'
epn1hXs.append(epn1_df.loc[hole1X] + offset1X)
epn1hYs.append(epn1_df.loc[hole1Y] + offset1Y)
epn1hZs.append(epn1_df.loc[hole1Z] + offset1Z)
# Moves the epn2 stack from its default (0,0,0) positions.
offset2X, offset2Y, offset2Z = -0.25, +0.25, +0.25
#for j in range (epn2_df.loc['Protons']):
for j in range (epn2protons):
# Randomize the dataframe's fixed
# electron and neutron coordinates.
aze2 = random()*PI_PHI
azn2 = random()*PI_PHI
epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]] = rotateAbtZ(epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]], aze2)
epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]] = rotateAbtZ(epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]], azn2)
if orient2 == '-z':
epn2_df.loc[proXColList[j]], epn2_df.loc[proYColList[j]], epn2_df.loc[proZColList[j]] = rotateAbtX(epn2_df.loc[proXColList[j]], epn2_df.loc[proYColList[j]], epn2_df.loc[proZColList[j]], PI_PHI)
epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]] = rotateAbtX(epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]], PI_PHI)
epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]] = rotateAbtX(epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]], PI_PHI)
epn2_df.loc['SlotOrien'] = '-Z'
elif orient2 == 'x':
epn2_df.loc[proXColList[j]], epn2_df.loc[proYColList[j]], epn2_df.loc[proZColList[j]] = rotateAbtY(epn2_df.loc[proXColList[j]], epn2_df.loc[proYColList[j]], epn2_df.loc[proZColList[j]], PI_PHI/2)
epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]] = rotateAbtY(epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]], PI_PHI/2)
epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]] = rotateAbtY(epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]], PI_PHI/2)
epn2_df.loc['SlotOrien'] = 'X'
elif orient2 == '-x':
epn2_df.loc[proXColList[j]], epn2_df.loc[proYColList[j]], epn2_df.loc[proZColList[j]] = rotateAbtY(epn2_df.loc[proXColList[j]], epn2_df.loc[proYColList[j]], epn2_df.loc[proZColList[j]], -PI_PHI/2)
epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]] = rotateAbtY(epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]], -PI_PHI/2)
epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]] = rotateAbtY(epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]], -PI_PHI/2)
epn2_df.loc['SlotOrien'] = '-X'
elif orient2 == 'y':
epn2_df.loc[proXColList[j]], epn2_df.loc[proYColList[j]], epn2_df.loc[proZColList[j]] = rotateAbtX(epn2_df.loc[proXColList[j]], epn2_df.loc[proYColList[j]], epn2_df.loc[proZColList[j]], -PI_PHI/2)
epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]] = rotateAbtX(epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]], -PI_PHI/2)
epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]] = rotateAbtX(epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]], -PI_PHI/2)
epn2_df.loc['SlotOrien'] = 'Y'
elif orient2 == '-y':
epn2_df.loc[proXColList[j]], epn2_df.loc[proYColList[j]], epn2_df.loc[proZColList[j]] = rotateAbtX(epn2_df.loc[proXColList[j]], epn2_df.loc[proYColList[j]], epn2_df.loc[proZColList[j]], PI_PHI/2)
epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]] = rotateAbtX(epn2_df.loc[eleXColList[j]], epn2_df.loc[eleYColList[j]], epn2_df.loc[eleZColList[j]], PI_PHI/2)
epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]] = rotateAbtX(epn2_df.loc[neuXColList[j]], epn2_df.loc[neuYColList[j]], epn2_df.loc[neuZColList[j]], PI_PHI/2)
epn2_df.loc['SlotOrien'] = '-Y'
else :
epn2_df.loc['SlotOrien'] = 'Z'
# All protons
#epn2AllpXs.append(epn2_df.loc[proXColList[j]] + offset2X)
epn2AllpXs.append(epn2_df.loc[proXColList[j]] + offset2X)
epn2AllpYs.append(epn2_df.loc[proYColList[j]] + offset2Y)
epn2AllpZs.append(epn2_df.loc[proZColList[j]] + offset2Z)
# Protons - Left or Right
if epn2_df.loc[proSpColList[j]] == 'L':
epn2pLXs.append(epn2_df.loc[proXColList[j]] + offset2X)
epn2pLYs.append(epn2_df.loc[proYColList[j]] + offset2Y)
epn2pLZs.append(epn2_df.loc[proZColList[j]] + offset2Z)
else :
epn2pRXs.append(epn2_df.loc[proXColList[j]] + offset2X)
epn2pRYs.append(epn2_df.loc[proYColList[j]] + offset2Y)
epn2pRZs.append(epn2_df.loc[proZColList[j]] + offset2Z)
# All Electrons
epn2AlleXs.append(epn2_df.loc[eleXColList[j]] + epn2_df.loc[proXColList[j]] + offset2X)
epn2AlleYs.append(epn2_df.loc[eleYColList[j]] + epn2_df.loc[proYColList[j]] + offset2Y)
epn2AlleZs.append(epn2_df.loc[eleZColList[j]] + epn2_df.loc[proZColList[j]] + offset2Z)
# Electrons - Top or Bottom
if epn2_df.loc[elePoColList[j]] == 'T':
epn2eTXs.append(epn2_df.loc[eleXColList[j]] + epn2_df.loc[proXColList[j]] + offset2X)
epn2eTYs.append(epn2_df.loc[eleYColList[j]] + epn2_df.loc[proYColList[j]] + offset2Y)
epn2eTZs.append(epn2_df.loc[eleZColList[j]] + epn2_df.loc[proZColList[j]] + offset2Z)
else :
epn2eBXs.append(epn2_df.loc[eleXColList[j]] + epn2_df.loc[proXColList[j]] + offset2X)
epn2eBYs.append(epn2_df.loc[eleYColList[j]] + epn2_df.loc[proYColList[j]] + offset2Y)
epn2eBZs.append(epn2_df.loc[eleZColList[j]] + epn2_df.loc[proZColList[j]] + offset2Z)
# Neutrons
epn2nXs.append(epn2_df.loc[neuXColList[j]] + epn2_df.loc[proXColList[j]] + offset2X)
epn2nYs.append(epn2_df.loc[neuYColList[j]] + epn2_df.loc[proYColList[j]] + offset2Y)
epn2nZs.append(epn2_df.loc[neuZColList[j]] + epn2_df.loc[proZColList[j]] + offset2Z)
# Hole
epn2hXs.append(epn2_df.loc[hole2X] + offset2X)
epn2hYs.append(epn2_df.loc[hole2Y] + offset2Y)
epn2hZs.append(epn2_df.loc[hole2Z] + offset2Z)
print("orient1 is ",orient1,". orient2 is ",orient2)
print('epn1: offset1X, offset1Y, offset1Z = ', offset1X, offset1Y, offset1Z)
print('epn2: offset2X, offset2Y, offset2Z = ', offset2X, offset2Y, offset2Z)
print('epnStacks[numberepn1-1] = ', epnStacks[numberepn1-1], 'epnStacks[numberepn2-1] = ', epnStacks[numberepn2-1])
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
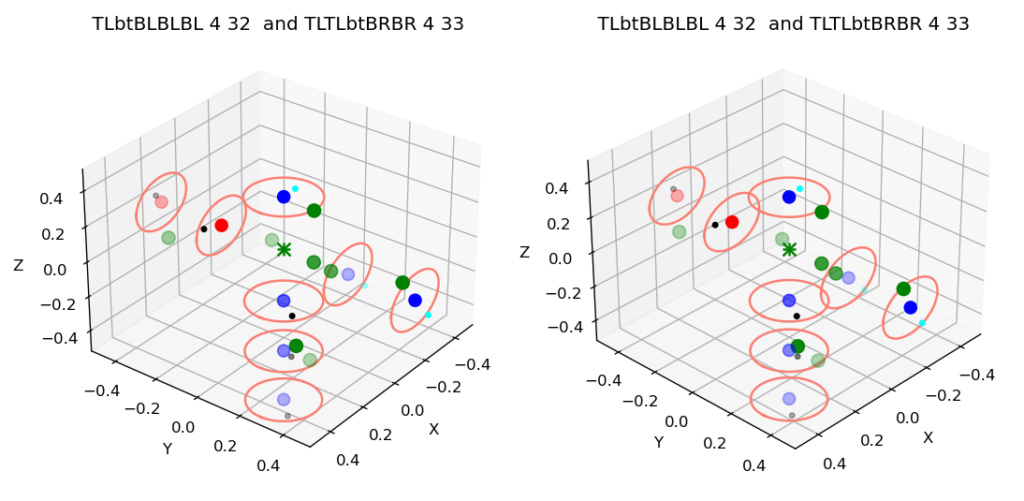
Happy New Year! If you cross your eyes a bit and stare, you should be able to see a single 3d image above.
Epn Bonding output cell (and plot) status.
Plot titles. As I’d mentioned - committed myself to last time, I was able to add the improved, coded formal Plot title to the three matplotlib plots: the two for plotting single atoms and third for plotting the two epn stacks. By using string conversions of tuples element[] and epnStacks[].
- Code:
# Creating the plot title.
t1, t2 = epnStacks[numberepn1-1][0],epnStacks[numberepn2-1][0]
title1 = ''.join(t1)
title2 = ''.join(t2)
plotTitle = title1 + ' and' + title2
Simple enough, another example of something I might have thought of sooner, I’m thankful for small progress.
FindHole(). And more satisfying progeress, added findHole(), a function that’s used to: 1. find each epn stacks' four hole indices – (h1-h7); (h1X,h1Y,h1Z), … , (h7X,h7Y,h7Z), rotates the hole’ coordinates to the desired direction and returns those values which are then copied to epn1 or epn2 dataframes. I noted that it is highly unreccommended to make changes to global variables (hole, holeX, holeY, holeZ ) from within a function, in this case, from what little I understand, I think it’s an acceptable solution. Now I need to functionalize the rest of the stack. At which time I believe it would be easy to add another epn stack to the plot.
ShowNeutrons. Added a ‘ShowNeutrons’ checkbox to the bond widget gui – to include neutrons in the plot or not.
Offsets. Still trying to figure out how to present bonding alternatives. In the image above, each epn stack’s center is offset to the other epn’s hole.
Bonding widget cell code. Cleaned up a bit with the findHole() function and removing an erroneous, redundant block of proton, neutron and electron rotations beginning with “ if orient1 == '-z': …”, ... . Anyway, the latest has 30 lines less code, to big to show with the two images, t’s been uploaded to github. https://github.com/LtAirman/ChargeFieldTopics
Then, by golly, the TRBL emissions appear to have been offset in the wrong direction -z instead of -y as in the image below. All four protons are at z = 0. I'll cross my fingers too and hope it’s easily remedied.

.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Here are two ‘eye crossing’ 3d charge field atomic models of Tellurium and Iron.
Plot changes. Corrected two (‘i’ to ‘m’) indices in the plot cell’s epn2 x emission section that clears-up my previous post’s second image problem – x-aligned stack with emissions in the wrong location.
Bond widget output cell changes. Added the round() function to all epn1-df and epn2_df hole and particle position float values.
Added two lists of 24 lists - epn1Lists, epn2Lists, for each epn stack’s hole and epn particle’s (x,y,z) coordinate positions. All positional amendments are now made with the appropriate list index.
Replaced the two rotate stack (epn1 and epn2) sections with a single orientStackEPNs() function.
The function parameters:
def orientStackEPNs(epn_df, epnLists, epnprotons, holeX, holeY, holeZ, orient, offsetX, offsetY, offsetZ):
The two function call arguments:
orientStackEPNs(epn1_df, epn1Lists, epn1protons, hole1X, hole1Y, hole1Z, orient1, offset1X, offset1Y, offset1Z)
orientStackEPNs(epn2_df, epn2Lists, epn2protons, hole2X, hole2Y, hole2Z, orient2, offset2X, offset2Y, offset2Z)
The bonding cell’s code is at 238 lines.
- Code:
"""
Bonding widget output under construction.
A Bonding widget will allow closer study to help
determine the validity of bonds between adjacent
orthogonal epn stacks.
An epn (electrons, protons and neutrons) stack
consists of 1-6 epn sets. Each stack includes a
low charge field pressure zone (low-cfz) 'hole'
at either the stack's top, bottom, or somewhere
in the middle. I'll assume bonding between two
stacks should occur at one or both stack holes.
Build selected epns.
"""
# These 12 lists includes the header 'loc's for all epn
# particles and holes' x y z coordinates.
# This series of 12 lists point to the columns
# containing up to 18 (x,y,z) particle coordinates
# and 7 (x,y,z) low-cfz hole coordinates.
# Added a proton spin col with values 'ps1',...,'ps6
# containing L or R proton spin and another row for
# each electron's T or B value, 'ep1',...,'ep6,
eleXColList = ['e1x', 'e2x', 'e3x', 'e4x', 'e5x', 'e6x']
eleYColList = ['e1y', 'e2y', 'e3y', 'e4y', 'e5y', 'e6y']
eleZColList = ['e1z', 'e2z', 'e3z', 'e4z', 'e5z', 'e6z']
elePoColList = ['ep1', 'ep2', 'ep3', 'ep4', 'ep5', 'ep6']
proXColList = ['p1x', 'p2x', 'p3x', 'p4x', 'p5x', 'p6x']
proYColList = ['p1y', 'p2y', 'p3y', 'p4y', 'p5y', 'p6y']
proZColList = ['p1z', 'p2z', 'p3z', 'p4z', 'p5z', 'p6z']
proSpColList = ['ps1', 'ps2', 'ps3', 'ps4', 'ps5', 'ps6']
neuXColList = ['n1x', 'n2x', 'n3x', 'n4x', 'n5x', 'n6x']
neuYColList = ['n1y', 'n2y', 'n3y', 'n4y', 'n5y', 'n6y']
neuZColList = ['n1z', 'n2z', 'n3z', 'n4z', 'n5z', 'n6z']
lowXColList = ['h1x', 'h2x', 'h3x', 'h4x', 'h5x', 'h6x', 'h7x']
lowYColList = ['h1y', 'h2y', 'h3y', 'h4y', 'h5y', 'h6y', 'h7y']
lowZColList = ['h1z', 'h2z', 'h3z', 'h4z', 'h5z', 'h6z', 'h7z']
# Initializing all the particle position lists.
# All protons in each epn stack
epn1AllpXs, epn1AllpYs, epn1AllpZs = [], [], []
epn2AllpXs, epn2AllpYs, epn2AllpZs = [], [], []
# Left spinning Protons.
epn1pLXs, epn1pLYs, epn1pLZs = [], [], []
epn2pLXs, epn2pLYs, epn2pLZs = [], [], []
# Right spinning Protons.
epn1pRXs, epn1pRYs, epn1pRZs = [], [], []
epn2pRXs, epn2pRYs, epn2pRZs = [], [], []
# All electrons
epn1AlleXs, epn1AlleYs, epn1AlleZs = [], [], []
epn2AlleXs, epn2AlleYs, epn2AlleZs = [], [], []
# Top proton pole Electrons.
epn1eTXs,epn1eTYs,epn1eTZs = [], [], []
epn2eTXs,epn2eTYs,epn2eTZs = [], [], []
# Bottom proton pole Electrons.
epn1eBXs,epn1eBYs,epn1eBZs = [], [], []
epn2eBXs,epn2eBYs,epn2eBZs = [], [], []
# Neutrons.
epn1nXs, epn1nYs, epn1nZs = [], [], []
epn2nXs, epn2nYs, epn2nZs = [], [], []
# Holes.
epn1hXs, epn1hYs, epn1hZs = [], [], []
epn2hXs, epn2hYs, epn2hZs = [], [], []
# The lists of each epn's lists and each list's index
epn1Lists = [epn1AllpXs, epn1AllpYs, epn1AllpZs, # 0,1,2
epn1pLXs, epn1pLYs, epn1pLZs, # 3,4,5
epn1pRXs, epn1pRYs, epn1pRZs, # 6,7,8
epn1AlleXs, epn1AlleYs, epn1AlleZs, # 9,10,11
epn1eTXs, epn1eTYs, epn1eTZs, # 12,13,14
epn1eBXs, epn1eBYs, epn1eBZs, # 15,16,17
epn1nXs, epn1nYs, epn1nZs, # 18,19,20
epn1hXs, epn1hYs, epn1hZs] # 21,22,23
epn2Lists = [epn2AllpXs, epn2AllpYs, epn2AllpZs, epn2pLXs, epn2pLYs, epn2pLZs,
epn2pRXs, epn2pRYs, epn2pRZs, epn2AlleXs, epn2AlleYs, epn2AlleZs,
epn2eTXs, epn2eTYs, epn2eTZs, epn2eBXs, epn2eBYs, epn2eBZs,
epn2nXs, epn2nYs, epn2nZs, epn2hXs, epn2hYs, epn2hZs]
#pd.options.display.max_columns = None
#pd.options.display.max_rows = None
# Copy the two selected epn rows from epnStacks to
# create single row dataframes epn1_df and epn2_df.
epn1_df = dfepn.loc[numberepn1-1, 'StackNumber':'e6z']
epn2_df = dfepn.loc[numberepn2-1, 'StackNumber':'e6z']
# Create a variable for the total number of protons
# in each stack.
epn1protons = epn1_df.loc['Protons']
epn2protons = epn2_df.loc['Protons']
#print('epn1_df',epn1_df,'epn2_df',epn2_df)
#print(epn1_df[:]) # Same output
# Find the correct (h1-h7) x,y,z coordinate column
# headers the data is in, and orient the hole.
def findHole(epn_df, orient):
global hole, holeX, holeY, holeZ
hole = epn_df.loc['Hloc']
holeX, holeY, holeZ = 'holeXW', 'holeYW', 'holeZW'
if hole == 'h1':
holeX, holeY, holeZ = 'h1x', 'h1y', 'h1z'
elif hole == 'h2':
holeX, holeY, holeZ = 'h2x', 'h2y', 'h2z'
elif hole == 'h3':
holeX, holeY, holeZ = 'h3x', 'h3y', 'h3z'
elif hole == 'h4':
holeX, holeY, holeZ = 'h4x', 'h4y', 'h4z'
elif hole == 'h5':
holeX, holeY, holeZ = 'h5x', 'h5y', 'h5z'
elif hole == 'h6':
holeX, holeY, holeZ = 'h6x', 'h6y', 'h6z'
elif hole == 'h7':
holeX, holeY, holeZ = 'h7x', 'h7y', 'h7z'
# Rotate the hole's initial +z orientation to the selected orientation.
if orient == '-z':
epn_df.loc[holeX], epn_df.loc[holeY], epn_df.loc[holeZ] = rotateAbtX(epn_df.loc[holeX], epn_df.loc[holeY], epn_df.loc[holeZ], PI_PHI)
elif orient == 'x':
epn_df.loc[holeX], epn_df.loc[holeY], epn_df.loc[holeZ] = rotateAbtY(epn_df.loc[holeX], epn_df.loc[holeY], epn_df.loc[holeZ], PI_PHI/2)
elif orient == '-x':
epn_df.loc[holeX], epn_df.loc[holeY], epn_df.loc[holeZ] = rotateAbtY(epn_df.loc[holeX], epn_df.loc[holeY], epn_df.loc[holeZ], -PI_PHI/2)
elif orient == 'y':
epn_df.loc[holeX], epn_df.loc[holeY], epn_df.loc[holeZ] = rotateAbtX(epn_df.loc[holeX], epn_df.loc[holeY], epn_df.loc[holeZ], -PI_PHI/2)
elif orient == '-y':
epn_df.loc[holeX], epn_df.loc[holeY], epn_df.loc[holeZ] = rotateAbtX(epn_df.loc[holeX], epn_df.loc[holeY], epn_df.loc[holeZ], PI_PHI/2)
epn_df.loc[holeX], epn_df.loc[holeY], epn_df.loc[holeZ] = round(epn_df.loc[holeX],2), round(epn_df.loc[holeY],2), round(epn_df.loc[holeZ],2)
return epn_df
# Globals that hold the hole index and coord location
hole, holeX, holeY, holeZ = 'unk', 'unk', 'unk', 'unk'
findHole(epn1_df, orient1)
# Copy the hole data
hole1, hole1X, hole1Y, hole1Z = hole, holeX, holeY, holeZ
print('Global hole1 = ',hole1,', hole1X = ', hole1X, ', hole1Y = ', hole1Y, ', hole1Z = ', hole1Z)
findHole(epn2_df, orient2)
hole2, hole2X, hole2Y, hole2Z = hole, holeX, holeY, holeZ
print('Global hole2 = ',hole2,', hole2X = ', hole2X, ', hole2Y = ', hole2Y, ', hole2Z = ', hole2Z)
# Offsets. Moves the epn1 stack from its default (0,0,0) positions
# Needed to move this section to after the findHole functions.
# A 'Not-working' erroneous flipped, y and Z emission locations
# were corrected in the plot's 2 'draw X emision' sections.
# Currently considering modifying the gui to include offets.
#offset1X, offset1Y, offset1Z = 0, 0, 0
#offset2X, offset2Y, offset2Z = 0, 0, 0
#offset1X, offset1Y, offset1Z = -0.25, -0.25, -0.25
#offset2X, offset2Y, offset2Z = 0.25, 0.25, 0.25
offset1X, offset1Y, offset1Z = -epn1_df.loc[hole1X], -epn1_df.loc[hole1Y], -epn1_df.loc[hole1Z]
offset2X, offset2Y, offset2Z = -epn2_df.loc[hole2X], -epn2_df.loc[hole2Y], -epn2_df.loc[hole2Z]
# Orient the stack particles.
def orientStackEPNs(epn_df, epnLists, epnprotons, holeX, holeY, holeZ, orient, offsetX, offsetY, offsetZ):
epnprotons = epn_df.loc['Protons']
for i in range (epnprotons):
# Rotate the stack, particle by particle, to the deired spin
# axis direction. epn1 dataframe's 'SlotOrien' value is updated.
# Randomize the dataframe's fixed electron and neutron coordinates.
aze = random()*PI_PHI
azn = random()*PI_PHI
epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]] = rotateAbtZ(epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]], aze)
epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]] = rotateAbtZ(epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]], azn)
if orient == '-z':
epn_df.loc[proXColList[i]], epn_df.loc[proYColList[i]], epn_df.loc[proZColList[i]] = rotateAbtX(epn_df.loc[proXColList[i]], epn_df.loc[proYColList[i]], epn_df.loc[proZColList[i]], PI_PHI)
epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]] = rotateAbtX(epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]], PI_PHI)
epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]] = rotateAbtX(epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]], PI_PHI)
epn_df.loc['SlotOrien'] = '-Z'
elif orient == 'x':
epn_df.loc[proXColList[i]], epn_df.loc[proYColList[i]], epn_df.loc[proZColList[i]] = rotateAbtY(epn_df.loc[proXColList[i]], epn_df.loc[proYColList[i]], epn_df.loc[proZColList[i]], PI_PHI/2)
epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]] = rotateAbtY(epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]], PI_PHI/2)
epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]] = rotateAbtY(epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]], PI_PHI/2)
epn_df.loc['SlotOrien'] = 'X'
elif orient == '-x':
epn_df.loc[proXColList[i]], epn_df.loc[proYColList[i]], epn_df.loc[proZColList[i]] = rotateAbtY(epn_df.loc[proXColList[i]], epn_df.loc[proYColList[i]], epn_df.loc[proZColList[i]], -PI_PHI/2)
epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]] = rotateAbtY(epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]], -PI_PHI/2)
epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]] = rotateAbtY(epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]], -PI_PHI/2)
epn_df.loc['SlotOrien'] = '-X'
elif orient == 'y':
epn_df.loc[proXColList[i]], epn_df.loc[proYColList[i]], epn_df.loc[proZColList[i]] = rotateAbtX(epn_df.loc[proXColList[i]], epn_df.loc[proYColList[i]], epn_df.loc[proZColList[i]], -PI_PHI/2)
epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]] = rotateAbtX(epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]], -PI_PHI/2)
epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]] = rotateAbtX(epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]], -PI_PHI/2)
epn_df.loc['SlotOrien'] = 'Y'
elif orient == '-y':
epn_df.loc[proXColList[i]], epn_df.loc[proYColList[i]], epn_df.loc[proZColList[i]] = rotateAbtX(epn_df.loc[proXColList[i]], epn_df.loc[proYColList[i]], epn_df.loc[proZColList[i]], PI_PHI/2)
epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]] = rotateAbtX(epn_df.loc[eleXColList[i]], epn_df.loc[eleYColList[i]], epn_df.loc[eleZColList[i]], PI_PHI/2)
epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]] = rotateAbtX(epn_df.loc[neuXColList[i]], epn_df.loc[neuYColList[i]], epn_df.loc[neuZColList[i]], PI_PHI/2)
epn_df.loc['SlotOrien'] = '-Y'
else :
epn_df.loc['SlotOrien'] = 'Z'
# Append the epn's particle type and coordinates to
# their respective five categorical lists for plotting.
# All protons
epnLists[0].append(round(epn_df.loc[proXColList[i]] + offsetX,2))
epnLists[1].append(round(epn_df.loc[proYColList[i]] + offsetY,2))
epnLists[2].append(round(epn_df.loc[proZColList[i]] + offsetZ,2))
# Protons - Left or Right
if epn_df.loc[proSpColList[i]] == 'L':
epnLists[3].append(round(epn_df.loc[proXColList[i]] + offsetX,2))
epnLists[4].append(round(epn_df.loc[proYColList[i]] + offsetY,2))
epnLists[5].append(round(epn_df.loc[proZColList[i]] + offsetZ,2))
else:
epnLists[6].append(round(epn_df.loc[proXColList[i]] + offsetX,2))
epnLists[7].append(round(epn_df.loc[proYColList[i]] + offsetY,2))
epnLists[8].append(round(epn_df.loc[proZColList[i]] + offsetZ,2))
# All Electrons
epnLists[9].append(round(epn_df.loc[eleXColList[i]] + epn_df.loc[proXColList[i]] + offsetX,2))
epnLists[10].append(round(epn_df.loc[eleYColList[i]] + epn_df.loc[proYColList[i]] + offsetY,2))
epnLists[11].append(round(epn_df.loc[eleZColList[i]] + epn_df.loc[proZColList[i]] + offsetZ,2))
# Electrons - Top or Bottom
if epn_df.loc[elePoColList[i]] == 'T':
epnLists[12].append(round(epn_df.loc[eleXColList[i]] + epn_df.loc[proXColList[i]] + offsetX,2))
epnLists[13].append(round(epn_df.loc[eleYColList[i]] + epn_df.loc[proYColList[i]] + offsetY,2))
epnLists[14].append(round(epn_df.loc[eleZColList[i]] + epn_df.loc[proZColList[i]] + offsetZ,2))
else :
epnLists[15].append(round(epn_df.loc[eleXColList[i]] + epn_df.loc[proXColList[i]] + offsetX,2))
epnLists[16].append(round(epn_df.loc[eleYColList[i]] + epn_df.loc[proYColList[i]] + offsetY,2))
epnLists[17].append(round(epn_df.loc[eleZColList[i]] + epn_df.loc[proZColList[i]] + offsetZ,2))
# Neutrons
epnLists[18].append(round(epn_df.loc[neuXColList[i]] + epn_df.loc[proXColList[i]] + offsetX,2))
epnLists[19].append(round(epn_df.loc[neuYColList[i]] + epn_df.loc[proYColList[i]] + offsetY,2))
epnLists[20].append(round(epn_df.loc[neuZColList[i]] + epn_df.loc[proZColList[i]] + offsetZ,2))
# Hole
epnLists[21].append(round(epn_df.loc[holeX] + offsetX,2))
epnLists[22].append(round(epn_df.loc[holeY] + offsetY,2))
epnLists[23].append(round(epn_df.loc[holeZ] + offsetZ,2))
return epn_df, epnLists, holeX, holeY, holeZ, orient
orientStackEPNs(epn1_df, epn1Lists, epn1protons, hole1X, hole1Y, hole1Z, orient1, offset1X, offset1Y, offset1Z)
orientStackEPNs(epn2_df, epn2Lists, epn2protons, hole2X, hole2Y, hole2Z, orient2, offset2X, offset2Y, offset2Z)
print("orient1 is ",orient1,". orient2 is ",orient2)
print('epn1: offset1X, offset1Y, offset1Z = ', offset1X, offset1Y, offset1Z)
print('epn2: offset2X, offset2Y, offset2Z = ', offset2X, offset2Y, offset2Z)
print('epnStacks[numberepn1-1] = ', epnStacks[numberepn1-1], 'epnStacks[numberepn2-1] = ', epnStacks[numberepn2-1])
Assuming the bonding rules also allow for single low-cf site bonding, my next goal is to make those alternative bonds user select-able.

P.S. Added Te and Fe slot layout diagrams.
.
Last edited by LongtimeAirman on Fri Jan 05, 2024 5:19 pm; edited 1 time in total (Reason for editing : Added P.S.)
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
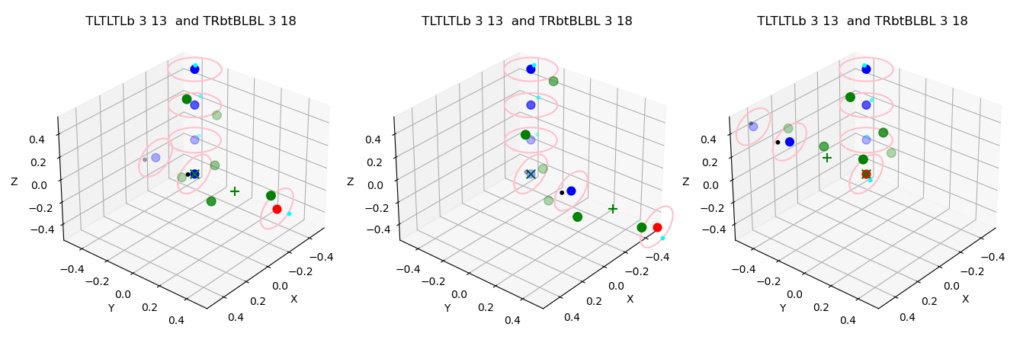
Don’t try crossing your eyes this time, these three different images all show the same z oriented epn stack TLTLTLb, with bottom hole bonding site (x) – ‘bound’ to each of the 3 protons (p1, p2, p3) of the y aligned epn stack TrbtBLBL.
Thanks Cr6. That helps me compensate for my lack of progress this week.
Adding a “stack Proton ‘bonding’ to an orthogonal stack hole” type bond. I started by building an interface in which the user could select the type of bond desired by selecting which of the two epn stacks contributes the bonding site hole with 'epn1', 'epn2', 'epn1&2'.
'epn1&2' Refers to the hole-to-hole bonding I’d previously shown. Basically, the ‘bond’ results by offsetting each epn’s stack (0,0,0) to its hole position. Done with the two lines:
- Code:
offset1X, offset1Y, offset1Z = -epn1_df.loc[hole1X], -epn1_df.loc[hole1Y], -epn1_df.loc[hole1Z]
offset2X, offset2Y, offset2Z = -epn2_df.loc[hole2X], -epn2_df.loc[hole2Y], -epn2_df.loc[hole2Z]
'epn1', 'epn2'. Selecting either of these two choices results in either epn1 or epn2 containing the desired bond hole. The proton that would end up at that hole is chosen by selecting ‘bondproton’ with possible proton values ‘p1’, ‘p2’, ‘p3’, ‘p4’, ‘p5’, ‘p6’.
- Code:
# To create the new proton-to-hole type bond image above, 'epn1' was chosen as the hole location.
offset1X, offset1Y, offset1Z = -epn1_df.loc[hole1X], -epn1_df.loc[hole1Y], -epn1_df.loc[hole1Z]
# Then epn2’s three proton offset positions were created separately with.
offset2X, offset2Y, offset2Z = 0, -epn2_df.loc[proZColList[0]], 0
#offset2X, offset2Y, offset2Z = 0, -epn2_df.loc[proZColList[1]], 0
#offset2X, offset2Y, offset2Z = 0, -epn2_df.loc[proZColList[2]], 0
I also added variables that limited the user’s ‘bondproton’ selection to the number of protons that stack actually contains, i.e. just p1, p2 or p3 for epn stack2 in the above image. I saw good plots at times and thought I could work with that, but more often than not I was plagued with plotting problems, such as missing or misplaced electrons, neutrons or emissions and scale changes with very elongated emissions. Very frustrating. I found that the ‘observed’ interactive gui variables often did not agree with the gui and plot settings. That led me to search for a possible solution, like
Python: Resetting A Matplotlib From An Ipywidget Button
https://stackoverflow.com/questions/43301231/python-resetting-a-matplotlib-from-an-ipywidget-button
Ended up commenting out all the gui changes except for 'epn1', 'epn2', 'epn1&2'.
Hope to make better progress this week.
P.S. Removed two redundant line of code.
,
Last edited by LongtimeAirman on Thu Jan 11, 2024 9:36 pm; edited 1 time in total (Reason for editing : Added P.S.)
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

After two stacks are chosen and oriented, if a proton-to-hole bond is desired, an interact widget can be used to select the stack proton that will occupy the other stack’s hole.
Here’s the code that created the output image shown.
- Code:
from ipywidgets import interact, Dropdown, Select
""" Select the Bonding Proton """
#if 1<= selectStack1.value-1 <= 4: epn1Ps = 1
#elif 5<= selectStack1.value-1 <= 12: epn1Ps = 2
#elif 13<= selectStack1.value-1 <= 24: epn1Ps = 3
#elif 25<= selectStack1.value-1 <= 40: epn1Ps = 4
#elif 41<= selectStack1.value-1 <= 60: epn1Ps = 5
#elif 61<= selectStack1.value-1 <= 84: epn1Ps = 6
epnPLocAllOptions = {
'One':['P1'],
'Two':['P1','P2'],
'three':['P1','P2','P3'],
'four':['P1','P2','P3','P4'],
'five':['P1','P2','P3','P4','P5'],
'six':['P1','P2','P3','P4','P5','P6']
}
epnProtonTot = Select(options = epnPLocAllOptions.keys())
bondProton = Select()
def update_bondProton_options(*args): # *args represent zero (case here) or more arguments.
bondProton.options = epnPLocAllOptions [epnProtonTot.value]
epnProtonTot.observe(update_bondProton_options)
@interact(epnPTot = epnProtonTot, pbond = bondProton)
def print_city(epnPTot, pbond):
print('Epn has',epnPTot,'proton and a bond at', pbond)
That code began as the suggested answer from a stack overflow question, https://stackoverflow.com/questions/28529157/dynamically-changing-dropdowns-in-ipython-notebook-widgets-and-spyre?rq=3 and my Brave browser AI.
- Code:
Me: How can I make a widget selection that depends on a previous widget selection using python ipywidgets?
AI: Sure! To get the value of a selected index type ipywidget, you can use the value property of the widget. Here is an example code snippet that demonstrates how to get the value of a Select widget:
import ipywidgets as widgets
# Create a list of options
options = ['Option 1', 'Option 2', 'Option 3']
# Create a Select widget
select_widget = widgets.Select(options=options)
# Display the widget
display(select_widget)
# Get the value of the selected option
selected_option = select_widget.value
In this example, selected_option will contain the value of the selected option. You can replace options with your own list of options and select_widget with your own Select widget.
I hope this helps! Let me know if you have any other questions.
Me: Is there a way to make 'option 1', 'option 2', and 'option 3' each present a different set of additional options?
Yes, you can create a dictionary of options where each key corresponds to an option in the Select widget and the value is a list of additional options. Here is an example code snippet that demonstrates how to do this:
import ipywidgets as widgets
# Create a dictionary of options
options = {
'Option 1': ['Option 1 - Additional Option 1', 'Option 1 - Additional Option 2'],
'Option 2': ['Option 2 - Additional Option 1', 'Option 2 - Additional Option 2'],
'Option 3': ['Option 3 - Additional Option 1', 'Option 3 - Additional Option 2']
}
# Create a Select widget
select_widget = widgets.Select(options=options.keys())
# Create a Dropdown widget for additional options
additional_options_widget = widgets.Dropdown()
# Define a function to update the additional options based on the selected option
def update_additional_options(*args):
additional_options_widget.options = options[select_widget.value]
# Register the update function to be called when the selected option changes
select_widget.observe(update_additional_options, 'value')
# Display the widgets
display(select_widget)
display(additional_options_widget)
In this example, options is a dictionary where each key corresponds to an option in the Select widget and the value is a list of additional options. The update_additional_options function updates the additional_options_widget based on the selected option in the select_widget. The observe method is used to register the update_additional_options function to be called when the selected option changes.
I hope this helps! Let me know if you have any other questions.
Being mainly interested in the interact and interactive ipywidgets, I’ve spent a good deal of time during the last week reviewing the first 52 minutes of a three hour video, The Jupyter Interactive Widget Ecosystem | Scipy 2019 Tutorial | Breddels, Craig, Grout, Renou https://www.youtube.com/watch?v=HaSpqsKaRbo
I downloaded the source files so I could work all the tutorial examples, noting that they included a few small changes for SciPy 2022. https://github.com/jupyter-widgets/tutorial?tab=readme-ov-file
Also reviewed several other ‘interact’ and ‘interactive’ videos and articles but SciPy 2019 was easily the best. Most other sites merely provide copied portions of SciPy’s Jupyter-widget tutorials.
Latest changes to github, https://github.com/LtAirman/ChargeFieldTopics include:
1. Added a selectEPNHole widget with values 'epn1hole', 'epn1&2holes', and 'epn2hole' that I talked about last time.
2. Corrected another couple of bond plot cell typos in the 'x5' and 'y5' lines that accounted for elongated emission errors. I must have an evil twin that keeps making typos when I’m not looking.
3. ‘Select the Bonding Proton’. Added and am working on an interact widget cell - intended to select the proton for a hole-proton type bond, the code and cell’s output .
4. Adding a new temporary cell just after the ‘Select the Bonding Proton’ for print debugging while working on the interact widget.
5. Moved the, ‘create single row dataframes epn1_df and epn2_df’, to its own cell just before the Bonding widget output cell.
I must admit I’ll probably need to do a lot more studying before I can comprehend the attached code. For example, rather than presenting the two dropdowns as shown, it should be possible to present a single list that contains only the protons in the selected stack.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
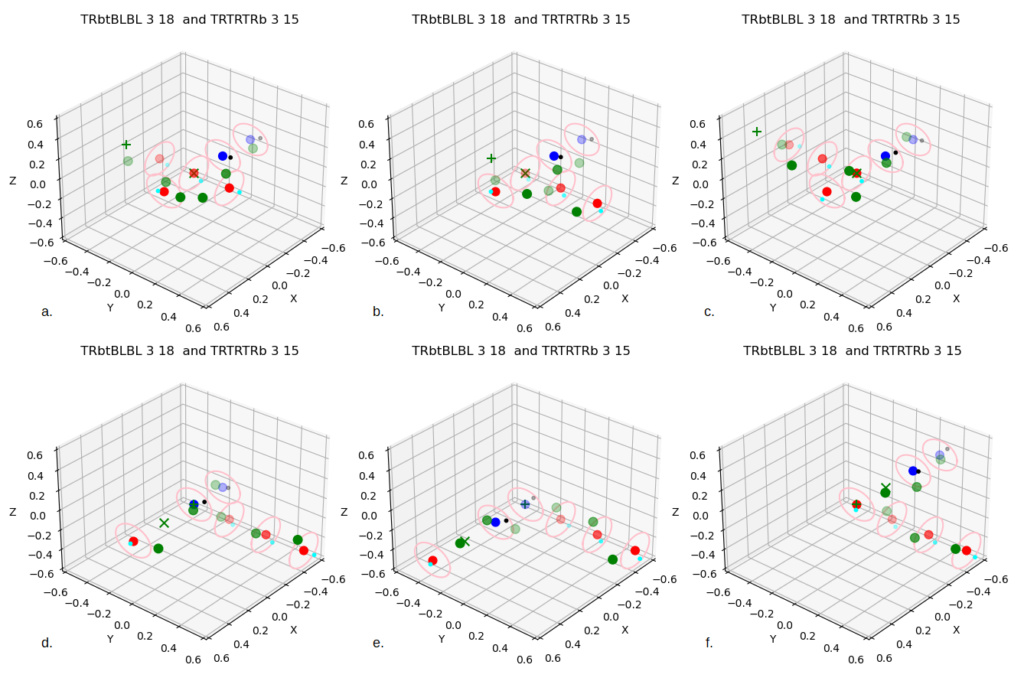
The bond widget can now display user selected orthogonal ‘proton-to-hole’ bonds, where any proton from epn2 can bond with epn1’s hole (x), or any epn1 proton can bond with epn2’s hole (+). In the top row, a b and c, the +x aligned epn1 stack TRbtBLBL provides the hole, and the +y aligned TRTRTRb is shown positioned with its three protons P1, P2 and P3 filling that hole. P1 is the center proton and P3 is the ‘top’ proton in both stacks. In the bottom row, d e and f, the +y aligned epn2 TRTRTRb provides the epn2 hole, and the +x aligned epn1 stack TRbtBLBL is shown with each of its protons filling that hole.
Of course we don’t know whether any or all of these so-called bonding configurations are valid or not. In my opinion, the utility of the bonding widget is in allowing a closer examination of the various possible hole-to-hole or proton-to-hole bonds in order to help identify the charge field bonding rules.
Next, the bonding widget should also include in-line, parallel or anti-parallel single stack bonds that can occur when the two epn stacks total six or less protons.
Any given epn stack contains a single low charge field pressure hole (electron vacant) either at one stack end or the other or at a bt alpha center somewhere in the middle. A hole is needed in any hole-to-hole or proton-to-hole bond. I’m beginning to wonder how to go about improving the atomic plots by including the holes as the primary molecular bonding sites.
I managed to limit the user selection to just the protons present with a single dropdown list, without the need for interact or interactive widgets, by creating the AI suggested ‘dictionary of options’. Here’s the current code, minus the commentary.
- Code:
""" Select the Bonding Proton """
# The 1-84 epnStacks[] are sorted by stack size, 1-6.
epnPs = 1 # Initializing epnPs
# The proton position options for 1-6 epn stacks.
epnPLocAllOptions = {
1 : ['P1'],
2 : ['P1','P2'],
3 : ['P3','P1','P2'],
4 : ['P3','P1','P2','P4'],
5 : ['P5','P3','P1','P2','P4'],
6 : ['P5','P3','P1','P2','P4','P6']}
if selectEPNHole.value == 'epn1hole' :
# Find epn2's proton.
if 1<= selectStack2.value <= 4: epnPs = 1
elif 5<= selectStack2.value <= 12: epnPs = 2
elif 13<= selectStack2.value <= 24: epnPs = 3
elif 25<= selectStack2.value <= 40: epnPs = 4
elif 41<= selectStack2.value <= 60: epnPs = 5
elif 61<= selectStack2.value <= 84: epnPs = 6
elif selectEPNHole.value == 'epn2hole' :
if 1<= selectStack1.value <= 4: epnPs = 1
elif 5<= selectStack1.value <= 12: epnPs = 2
elif 13<= selectStack1.value <= 24: epnPs = 3
elif 25<= selectStack1.value <= 40: epnPs = 4
elif 41<= selectStack1.value <= 60: epnPs = 5
elif 61<= selectStack1.value <= 84: epnPs = 6
elif selectEPNHole.value == 'epn1&2holes' :
# Hole-to-hole bonds do not include a proton
epnPLocAllOptions = {
1 : ['zero'],
2 : ['zero'],
3 : ['zero'],
4 : ['zero'],
5 : ['zero'],
6 : ['zero']}
protonSelect = widgets.Dropdown(
options= epnPLocAllOptions[epnPs],
#value='none', # The default value
description='ProtonBond',)
protonSelect
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
https://www.youtube.com/watch?v=bXrrVddn4Qw
https://github.com/BrainJS/brain.js
Might be cool to create a Co-Pilot for Miles' site: https://www.youtube.com/watch?v=pLiIK2kryw0
Lately I've been working with AuraDB (Neo4j) to build a graph database of Miles' work. Was looking at the following to get this together. LLMs with GraphDBs looks like an interesting future. Not totally up to speed with Cypher queries. Pretty much abandoned Tinkerpop-OrientDB due to limitations. If we create a graph database for the Miles' Charge Field it will likely scale better with Neo4j-AuraDB...at least this is my thinking. Currently with the Node-Relationship...it is tricky to say this is related to that when layout geometry is involved. Basically, I'm currently trying to port the SQL Physics database over to AuraDB. Importing into AuraDB is still trickier than it should be. I may have export everything in T-SQL queries for the GraphDB in AuraDB. https://neo4j.com/docs/cypher-cheat-sheet/5/auradb-enterprise/#_path_patterns Sorry LTAM and Lloyd...this is kind of amateur "hour" for me to get this stood up and working. AuraDB can scale...so if we get an algo that can show C.F. relationships via Graph from Atoms to Molecules via C.F. bonding rules...it should be easier and more flexible:
https://neo4j.com/docs/cypher-cheat-sheet/5/auradb-enterprise/#_path_patterns
The "relationships" between atoms is challenge. Sadly, Aura DB just does .csv imports and not SQL Server connections. May need to create a set of files to be imported for nodes-relationships. I think it can be done. AuraDB is pretty flexible for modeling Miles' structures for queries and even generative queries. AI-ChatGPT can help show off Miles' structures IMHO.
https://neo4j.com/blog/unifying-llm-knowledge-graph/
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
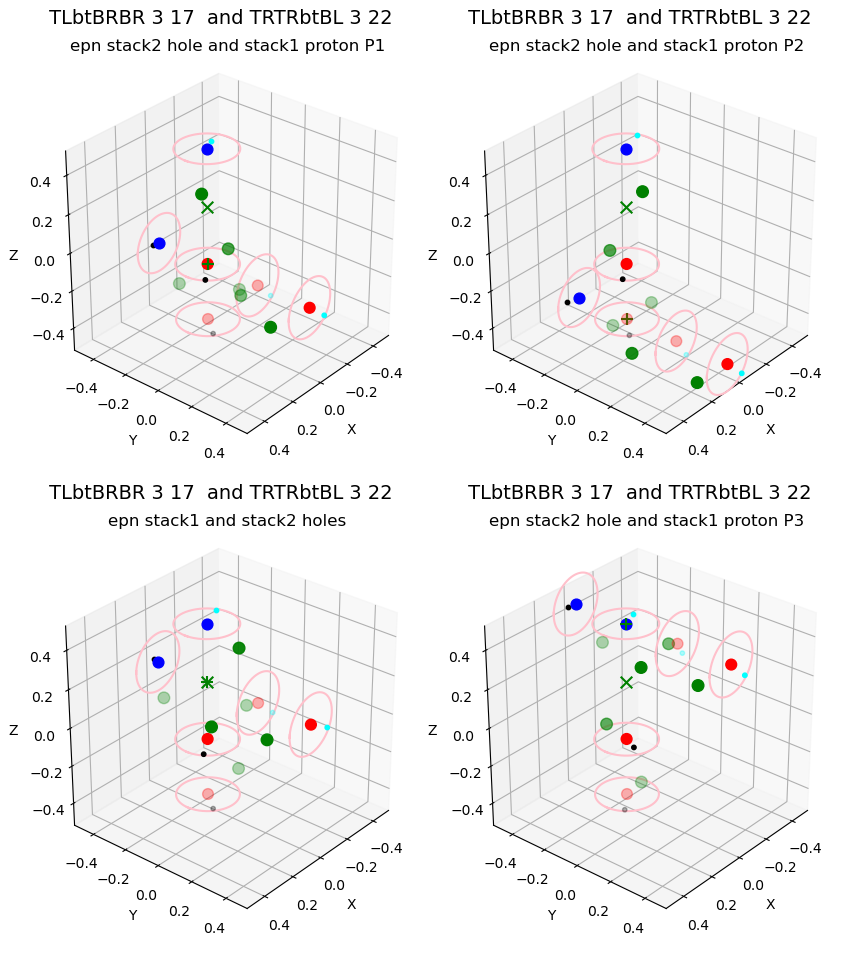
1. The image shows 4 separate plots. Changed each plot’s ax.set_title(‘plotTitle’), in this case ‘TLbtBRBR 3 17 and TRTRbtBL 3 22’, to a fig.suptitle(‘plotTitle’); then added a second title bond description -‘epn stack2 hole and stack1 proton P1’ to ax.set_title(bondmsg).
2. Added a code cell, about 130 lines including:
offsetx = x1max + (x1max-x1min)/2 + x2max + (x2max-x2min)/2
to offset both epn stacks’ centers such that both stacks’ particles and holes are geometrically centered to fit the plot’s +/- x y and z extents.
3. Added a plot command ax.set_box_aspect((1, 1, 1)) in order make the z axis scale match the x and y scales, unflattening the previous ‘ottoman-like’ carousel alpha center.
Haven’t made any progress in adding in-line single-stack parallel and anti-parallel bonds to the bond widget. Or in coming up with any way to validate bonds.
Cr6 I looked at your AI references and more. Optimizing the notebook is a good idea. So is creating a Co-Pilot for Miles’ site. I’m weakening. My resistance is futile. Computer science is experiencing rapid changes. CS majors will be expected to master multiple Large Language and AI Coding Assistant alternatives. Personally, I kinda like Github Copilot, even if it does cost ten dollars a month.
Your ideas are bigger than mine amigo, I’d be happy to follow your lead.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
https://github.com/mindsdb/mindsdb/tree/staging
Features available.
-------------
With this way we may be able to create a Knowledge Graph for Miles' stuff and upload the Graph DB model to an AuraDB which can scale. Then use this Neo4j LLM for creating Cypher queries -- https://neo4j.com/blog/unifying-llm-knowledge-graph/
GenAI-Stack (docker file ) (only runs well on Linux or Apple)
https://github.com/docker/genai-stack
There's a cool feature that let's you add .pdf text from .pdf files to a model so that It can be queried in Natural Language.
Trying to get this going at the moment. I'm going to use Graph DBs from AuraDB-Neo4j since they can scale. Not looking at other solutions now like OrientDB-Tinkerpop-Apache Age which I have working since they all require a lot of extra database configuration in Linux.
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
In Diatomic Hydrogen, Miles describes all the charge field rules needed to bond 2-6 ep (electron/proton) pairs into a single stack. For a quick review (each iteration helps improve my understanding), vertically oriented protons (for reference) may spin right or left, R or L, red or blue above, and will be accompanied by a single electron positioned at either the proton’s top or bottom pole, T or B, cyan or black as shown. Direct through charge flows between a proton’s poles from top to bottom or bottom to top. An R or L proton with T or B electron blocking one of the proton’ charge intakes creates a low charge field pressure hole near the proton’s opposite, vacant pole, enabling proton-to-proton bonding at that location. Protons without accompanying electrons cannot form a low charge pressure hole and so cannot bond. Ep-to-ep bonding with TT or BB electrons are valid charge field configurations. TB, an alpha type configuration is ok too, but vertically adjacent BT electrons, two adjacent protons with two electrons orbiting the proton poles between them interferes with and negates the low charge field zone formation, preventing the protons from maintaining their close positions and so cannot form a bond.
Another couple of small changes to the plot’s 2nd title bond description - ‘bondmsg’: 1. stack1, like the title above it is on the left, and stack 2 is on the right; 2. both sides, epn1 and epn2 now include the stacks’ +/- x y z orientation.
An entire stack’s set of ep’s may all spin left or right, or the stack may spin in two directions with a change in the middle, but the stack can only support a single change in the proton group’s spin direction and/or electron position. Such parallel (RR, LL) or anti-parallel (RL, LR) ep bonding results in a single shared direct charge field channel between and through all the stack’s protons. The charge passing through that channel is cumulative and increases with each added ep. A maximum stack of six ep’s can occur; however, a seventh ep cannot fill the 6-stack’s hole in parallel or anti-parallel position, the seventh ep’s will be turned by the overwhelming charge channel current into an orthogonal position with its emissions directed toward the 6-stack, thereby reducing the main charge it receives, allowing it to remain in the 6-stack’s low charge field hole. If an over current can turn the seventh proton, there might be an occasion where the seventh proton might actually cause a break somewhere in the middle the 6-stack. In the real world, atomic particles are subject to all sorts of prevailing charge field conditions, like the bond angle resulting between H2O atoms. I’m convinced that protons within an atom may increase in size sequentially, according to the atom’s ionization energy level, but that’s just a future possibility for now.
Miles’ charge field atomic emission diagram color coding, indicating the number of protons the stack contains remains the same. The colors are easily changeable. I’ve added colors and markers to differentiate R or L protons, T or B electrons, a neutron and a hole. In the TBRL code, the unfilled stack hole may be b or t or bt, small letter bottom or top or alpha center holes, located in one of up to 7 possible stack position locations adjacent to or between protons. The scale is arbitrary and the scatter point particle sizes vary. I’ve asserted that neutrons orbit the electron vacant proton pole and thereby enhance the stack’s low charge pressure hole, but otherwise, I don’t believe the neutrons are properly accounted for location wise just yet and can be omitted if desired. I would like the code to alter the distances between protons in the atomic plots, as Miles has recently described. I haven’t quite grasped the straightforward ratio Miles used to determine those distances, diminishing with increased number of protons, usually making the z-axis scale more compact than the lower proton count east-west and front-back channels. Haven’t decided whether or not to add charge channels to the bond plot, some combination of straight line direct charge channels through the protons’ spin axis, along with slender spirals and tiny tapered spirals indicating charge entering the electron vacant proton poles. It’d be nice if it were all animated.

The 84 epn stacks included in the widget are, to the best of my understanding, all the possible valid, parallel and anti-parallel proton stack configurations that can exist. The bond widget can display any pair of 84X84 epn stack combinations arranged in any 1 of 24 possible orthogonal orientations.
Now that we can ‘closely review’ possible bonds between epn stacks, how do we go about determining whether any given bond is valid or not? For the time being, I’m afraid we’re limited to discussion. For example, what can be said for about:
1. x aligned TLTLbtBRBR stack1 hole and -y aligned TRTRbtBLBL stack2 hole.
2. x aligned TLTLbtBRBR stack1 hole and -y aligned TRTRbtBLBL stack2 proton P1?
1. The double alpha bond on the left looks fine to me.
2. I believe the possible bond stack2 P1 at stack1’s hole has problems. First, its lacking spin balance. Also P1 looks unstable, since it is subject to two opposite main charge channels at the same time toward its equator, it can be pushed sideways out of position along its spin axis. P1 in that position also seems to violate the ‘there can be only one change in electron position and/or proton spin direction in a given stack' rule, P1’s position in the resulting line of 5 protons involve two spin direction changes and two electron position changes. My guess is that the right side hole/P1 plot is not a valid bond.
Cr6, thanks for the status, direction and references, I’ll start trying to learn more about it.
*
http://milesmathis.com/index.html
332. Diatomic Hydrogen. My new charge bonding explains this much better than electron sharing. Plus an analysis of spin isomers. 9pp. http://milesmathis.com/diatom.pdf
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
https://www.youtube.com/watch?v=ftlZ0oeXYRE
https://www.youtube.com/watch?v=CEaDSOh_AoM
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Proof of life only – the placement of the two intersecting alpha, proton charge intakes are not yet correct; the two spirals at (-0.3,0,0) and (0,-0.3,0) are at the correct proton pole - opposite the electron, the closer two are not. Only one of either proton pairs shows the correct R or L spiraling direction. It might help explain how we may safely assume(?) the bonding rules should include the Right hand rule.
Please pardon the delay. The usual reasons plus there was a storm and outage. I’m studying for a test. Paying close attention to the news, the occasional monster series and doom scrolling.
Charge channel objects. Tried adding small tapering proton pole spiral charge intakes. While I believe it is true that any given stack has only a single unfilled, available low charge pressure hole – unless that hole is filled with a lone proton without an attendant electron, there’s a hole associated with each and every proton in the plots shown.
Cr6, I’ve viewed the LLM and Neo4j references and am well-neigh overwhelmed. I may need to beg for more explicit directions. Will this stuff work on the notebook?
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Cr6, I’ve viewed the LLM and Neo4j references and am well-neigh overwhelmed. I may need to beg for more explicit directions. Will this stuff work on the notebook?
Yes, Neo4j does have a pip library for connections.
There are a few libraries out there for Neo4j.
https://neo4j.com/developer/python/
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Installed Neo4j using Anaconda, which indicates 5.17.0 is now present in my working environment.
Tried running the HelloWorldExample from
https://neo4j.com/docs/getting-started/languages-guides/neo4j-python/#python-driver
and received a service unavailable message included below.
- Code:
from neo4j import GraphDatabase
class HelloWorldExample:
def __init__(self, uri, user, password):
self.driver = GraphDatabase.driver(uri, auth=(user, password))
def close(self):
self.driver.close()
def print_greeting(self, message):
with self.driver.session() as session:
greeting = session.execute_write(self._create_and_return_greeting, message)
print(greeting)
@staticmethod
def _create_and_return_greeting(tx, message):
result = tx.run("CREATE (a:Greeting) "
"SET a.message = $message "
"RETURN a.message + ', from node ' + id(a)", message=message)
return result.single()[0]
if __name__ == "__main__":
greeter = HelloWorldExample("bolt://localhost:7687", "neo4j", "password")
greeter.print_greeting("hello, world")
greeter.close()
Output:
ServiceUnavailable: Couldn't connect to localhost:7687 (resolved to ()):
Failed to establish connection to ResolvedIPv6Address(('::1', 7687, 0, 0)) (reason [WinError 10061] No connection could be made because the target machine actively refused it)
Failed to establish connection to ResolvedIPv4Address(('127.0.0.1', 7687)) (reason [WinError 10061] No connection could be made because the target machine actively refused it)
???
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Bond widget plots, now with (or without) the main proton charge intake spirals, rotated 90 degrees about z, to show the spiral ends.
Adding the charge intake spirals took a lot more coding than I’d anticipated, starting with 24 new lists for the appropriate proton hole coordinates, opposite the electron, for the 2 stacks.
1. Left spin, top hole protons. epn1pLTXs[], epn1LTYs[], epn1LTZs[].
2. Left spin, top hole protons. epn2pLTXs[], epn2LTYs[], epn2LTZs[].
3. Left spin, bottom hole protons. epn1pLBXs[], epn1LBYs[], epn1LBZs[].
4. Left spin, bottom hole protons. epn2pLBXs[], epn2LBYs[], epn2LBZs[].
5. Right spin, top hole protons. epn1pRTXs[], epn1RTYs[], epn1RTZs[].
6. Right spin, top hole protons. epn2pRTXs[], epn2RTYs[], epn2RTZs[].
7. Right spin, bottom hole protons. epn1pRTXs[], epn1RTYs[], epn1RTZs[].
8. Right spin, bottom hole protons. epn2pRTXs[], epn2RTYs[], epn2RTZs[].
Appending each proton location to its appropriate set of lists added 40 lines to the ‘Bonding Widget Output’ cell.
Stationary R spinning protons have R spinning spirals and L spinning protons have L spinning spirals. Each stack needs to be aligned to the desired +/- x, y and z directions, adding 300 new code lines to the plot cell. The first two of the eight spiral plotting sections are as follows:
- Code:
cycles = 4
springR = 0.125
#cem = 'lightgrey'
cemr = 'pink'
ceml = 'lightblue'
cem = 'silver'
ts = np.arange(0, cycles*2*np.pi+.1, 0.01)
# epn1pLTXs. Left spin, top hole protons. epn1pLTXs, epn1LTYs, epn1LTZs,
for k in range(len(epn1pLTXs)):
if (orient1 == 'z') or (orient1 =='-z'):
if orient1 == 'z': #
xL1 = epn1pLTXs[k] + 0.05*ts*springR*np.cos(-ts)
yL1 = epn1pLTYs[k] + 0.05*ts*springR*np.sin(-ts)
zsL1 = epn1pLTZs[k] + ts*0.005
ax.plot(xL1, yL1, zsL1, zdir='z', c=ceml) # z
elif orient1 == '-z':
xR1 = epn1pLTXs[k] - 0.05*ts*springR*np.cos(ts)
yR1 = epn1pLTYs[k] - 0.05*ts*springR*np.sin(ts)
zsR1 = epn1pLTZs[k] - ts*0.005
ax.plot(xR1, yR1, zsR1, zdir='z', c=ceml) # -z
elif (orient1 == 'x') or (orient1 =='-x'):
if orient1 == 'x' :
xs2 = epn1pLTYs[k] + 0.05*ts*springR*np.cos(-ts)
ys2 = epn1pLTZs[k] + 0.05*ts*springR*np.sin(-ts)
zss2 = epn1pLTXs[k] + ts*0.005
ax.plot(xs2, ys2, zss2, zdir='x', c=ceml) # x
elif orient1 =='-x':
xs2 = epn1pLTYs[k] - 0.05*ts*springR*np.cos(ts)
ys2 = epn1pLTZs[k] - 0.05*ts*springR*np.sin(ts)
zss2 = epn1pLTXs[k] - ts*0.005
ax.plot(xs2, ys2, zss2, zdir='x', c=ceml) # -x
elif (orient1 == 'y') or (orient1 =='-y'):
if orient1 == 'y' :
xs3 = epn1pLTXs[k] + 0.05*ts*springR*np.cos(ts)
ys3 = epn1pLTZs[k] + 0.05*ts*springR*np.sin(ts)
zss3 = epn1pLTYs[k] + ts*0.005
ax.plot(xs3, ys3, zss3, zdir='y', c=ceml) # y
elif orient1 == '-y' :
xs3 = epn1pLTXs[k] - 0.05*ts*springR*np.cos(-ts)
ys3 = epn1pLTZs[k] - 0.05*ts*springR*np.sin(-ts)
zss3 = epn1pLTYs[k] - ts*0.005
ax.plot(xs3, ys3, zss3, zdir='y', c=ceml) #-y
# epn2pLTXs. Left spin, top hole protons. epn2pLTXs, epn2LTYs, epn2LTZs,
for k in range(len(epn2pLTXs)):
if (orient2 == 'z') or (orient2 =='-z'):
if orient2 == 'z': #
xL1 = epn2pLTXs[k] + 0.05*ts*springR*np.cos(-ts)
yL1 = epn2pLTYs[k] + 0.05*ts*springR*np.sin(-ts)
zsL1 = epn2pLTZs[k] + ts*0.005
ax.plot(xL1, yL1, zsL1, zdir='z', c=ceml) # z
elif orient2 == '-z':
xR1 = epn2pLTXs[k] - 0.05*ts*springR*np.cos(ts)
yR1 = epn2pLTYs[k] - 0.05*ts*springR*np.sin(ts)
zsR1 = epn2pLTZs[k] - ts*0.005
ax.plot(xR1, yR1, zsR1, zdir='z', c=ceml) # -z
elif (orient2 == 'x') or (orient2 =='-x'):
if orient2 == 'x' :
xs2 = epn2pLTYs[k] + 0.05*ts*springR*np.cos(-ts)
ys2 = epn2pLTZs[k] + 0.05*ts*springR*np.sin(-ts)
zss2 = epn2pLTXs[k] + ts*0.005
ax.plot(xs2, ys2, zss2, zdir='x', c=ceml) # x
elif orient2 =='-x':
xs2 = epn2pLTYs[k] - 0.05*ts*springR*np.cos(ts)
ys2 = epn2pLTZs[k] - 0.05*ts*springR*np.sin(ts)
zss2 = epn2pLTXs[k] - ts*0.005
ax.plot(xs2, ys2, zss2, zdir='x', c=ceml) # -x
elif (orient2 == 'y') or (orient2 =='-y'):
if orient2 == 'y' :
xs3 = epn2pLTXs[k] + 0.05*ts*springR*np.cos(ts)
ys3 = epn2pLTZs[k] + 0.05*ts*springR*np.sin(ts)
zss3 = epn2pLTYs[k] + ts*0.005
ax.plot(xs3, ys3, zss3, zdir='y', c=ceml) # y
elif orient2 == '-y' :
xs3 = epn2pLTXs[k] - 0.05*ts*springR*np.cos(-ts)
ys3 = epn2pLTZs[k] - 0.05*ts*springR*np.sin(-ts)
zss3 = epn2pLTYs[k] - ts*0.005
ax.plot(xs3, ys3, zss3, zdir='y', c=ceml) # -y
These one sided spirals look a lot like the conic emissions I included and raved on about with the project’s earlier threejs atomic renderings. Perhaps these intakes ensure proton-to-proton particle separation? The spiral size, angle, number of rotations and length can be easily adjusted in the plot cell. I might add those control variables to the bonding widget. At present, the neutron and electron positions would need to be changed in EPNStacks.csv.
Concerning my Neo4j ‘HelloWorldExample’ WinError 10061.
Please pardon me, I’m no doubt at fault.
First for not having “created an instance”
https://neo4j.com/docs/python-manual/current/install/#get-an-instance
or connecting to the database.
https://neo4j.com/docs/python-manual/current/connect/
https://neo4j.com/docs/getting-started/
They offer some free on-line training.
https://graphacademy.neo4j.com/?ref=docs
I’ll try doing better at reading and following instructions.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
All the bonding widget changes are uploaded to github.
I see you’re providing a few operating instructions. With Neo4j Desktop? Will that interface with Jupyter Notebook? Rather than going off on any unnecessary tangents, and avoiding high anxiety, I would definitely appreciate your approval/confirmation before proceeding with a Neo4j Desktop install.Cr6 wrote. Neo4j Desktop can throw some curve balls. …
Starting at https://neo4j.com/developer/graph-database/
after the quick 1 minute 18 second video, “Why Neo4J” four follow-up videos are displayed, one of which is, Neo4j (Graph Database) Crash Course
https://www.youtube.com/watch?v=8jNPelugC2s
by Laith Academy. In order to get started, Laith recommends that I download Neo4j Desktop from https://neo4j.com/download/
The Nov 29, 2021 video shows version 4.3 but the link now shows the current version, Neo4j Enterprise 5.16.0 for Developers. The requirement for Windows is 8.1+ w/Powershell, I have Windows 10 w/Powershell.
About 10 minutes into the 1 hour and 23 minute video Laith shows us the downloading process. He fills out an initial form, then clicks download. The download begins with a Neo4j page containing an authentication key and links to an installation guide and video. After neo4j is downloaded, Laith clicks on it. He’s using a Mac operating system, so he drags neo4j desktop to the application folder. He mentions that installing Neo4j Desktop on a windows system involves a wizard and is very similar. Next one needs to click run. The license is presented, he agrees with it, and is then redirected to the software registration where he pastes a copy of the authentication key. He is then able to activate the account and launch Neo4j Desktop. His tutorial uses NBA basketball database that can be found at a github link https://github.com/harblaith7/Neo4j-Crash-Course/blob/main/01-initial-data.cypher.
Do I understand correctly? If this is what you have in mind, I’ll try downloading Neo4j Desktop and completing the video. If not, I await further instructions.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference
Sample notebook: https://github.com/tomasonjo/blogs/blob/master/gds_python/gds_python_intro.ipynb
Samples of gds :
https://github.com/neo4j/graph-data-science-client/blob/main/examples/load-data-via-graph-construction.ipynb
https://neo4j.com/docs/graph-data-science/2.6/getting-started/fastrp-knn-example/
Chromium6- Posts : 727
Join date : 2019-11-29
 Re: Miles Periodic Table with Standard Periodic Table reference
Re: Miles Periodic Table with Standard Periodic Table reference

Good news and bad news.
Good news. I registered to enroll in the ‘courses’ at GraphAcademy, https://graphacademy.neo4j.com/
and have completed three so far. 1. Neo4j Fundamentals; 2. Cypher Fundamentals; and 3. Graph Data Modeling Fundamentals.
The courses use the neo4j sandbox in the clouds, I also downloaded neo4J Desktop to my machine.
Bad News. When I tried loading the Movie Graph project included in the install, I receive the neo4j Browser errors: ‘Connection failed could not connect…’, and ‘ERROR BOLTConnectionError. No connection found, did you connect to neo4j?’
I see the first 3 (the third being - looking for neo4j ) of five initialization vertical circles checked, but not the fourth or fifth - it happens too quick to print the screen.
Don’t know what the problem is or what I’d need to do to correct it. Maybe if I followed your advice more closely(?).
I’ll take more course tutorials. Maybe there’s one on that 'connection' subject that could help. Or I might search for an applicable video somewhere else. Or ask for help from the 'neo4j 'community'. I may need to delete and reinstalling neo4j desktop.
.
LongtimeAirman- Admin
- Posts : 2027
Join date : 2014-08-10
Chromium6 likes this post
Page 10 of 12 •  1, 2, 3 ... 9, 10, 11, 12
1, 2, 3 ... 9, 10, 11, 12 
 Similar topics
Similar topics» Cr6's Periodic Table
» RSC's Periodic Table for Comparison
» Happy sesquicentennial to the periodic table of the elements
» Periodic Table of Toxic Elements